CroSR391 , an ortholog of the λ Cro repressor, plays a major role in suppressing polVR391 -dependent mutagenesis
- PMID: 34184328
- PMCID: PMC8460599
- DOI: 10.1111/mmi.14777
CroSR391 , an ortholog of the λ Cro repressor, plays a major role in suppressing polVR391 -dependent mutagenesis
Abstract
When subcloned into low-copy-number expression vectors, rumAB, encoding polVR391 (RumA'2 B), is best characterized as a potent mutator giving rise to high levels of spontaneous mutagenesis in vivo. This is in dramatic contrast to the poorly mutable phenotype when polVR391 is expressed from the native 88.5 kb R391, suggesting that R391 expresses cis-acting factors that suppress the expression and/or the activity of polVR391 . Indeed, we recently discovered that SetRR391 , an ortholog of λ cI repressor, is a transcriptional repressor of rumAB. Here, we report that CroSR391 , an ortholog of λ Cro, also serves as a potent transcriptional repressor of rumAB. Levels of RumA are dependent upon an interplay between SetRR391 and CroSR391 , with the greatest reduction of RumA protein levels observed in the absence of SetRR391 and the presence of CroSR391 . Under these conditions, CroSR391 completely abolishes the high levels of mutagenesis promoted by polVR391 expressed from low-copy-number plasmids. Furthermore, deletion of croSR391 on the native R391 results in a dramatic increase in mutagenesis, indicating that CroSR391 plays a major role in suppressing polVR391 mutagenesis in vivo. Inactivating mutations in CroSR391 therefore have the distinct possibility of increasing cellular mutagenesis that could lead to the evolution of antibiotic resistance of pathogenic bacteria harboring R391.
Keywords: DNA polymerase V; R391; SOS response; integrating conjugative element; mutagenesis.
© 2021 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Conflict of interest statement
The authors declare that they have no conflict of interest with the content of this article.
Figures
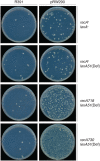






Similar articles
-
SetRICE391, a negative transcriptional regulator of the integrating conjugative element 391 mutagenic response.DNA Repair (Amst). 2019 Jan;73:99-109. doi: 10.1016/j.dnarep.2018.11.007. Epub 2018 Nov 16. DNA Repair (Amst). 2019. PMID: 30581075 Free PMC article.
-
Characterization of polVR391: a Y-family polymerase encoded by rumA'B from the IncJ conjugative transposon, R391.Mol Microbiol. 2007 Feb;63(3):797-810. doi: 10.1111/j.1365-2958.2006.05561.x. Mol Microbiol. 2007. PMID: 17302804
-
A λ Cro-Like Repressor Is Essential for the Induction of Conjugative Transfer of SXT/R391 Elements in Response to DNA Damage.J Bacteriol. 2015 Dec;197(24):3822-33. doi: 10.1128/JB.00638-15. Epub 2015 Oct 5. J Bacteriol. 2015. PMID: 26438816 Free PMC article.
-
Yet another way that phage λ manipulates its Escherichia coli host: λrexB is involved in the lysogenic-lytic switch.Mol Microbiol. 2015 May;96(4):689-93. doi: 10.1111/mmi.12969. Epub 2015 Mar 16. Mol Microbiol. 2015. PMID: 25684601 Review.
-
Insights into the complex levels of regulation imposed on Escherichia coli DNA polymerase V.DNA Repair (Amst). 2016 Aug;44:42-50. doi: 10.1016/j.dnarep.2016.05.005. Epub 2016 May 13. DNA Repair (Amst). 2016. PMID: 27236212 Free PMC article. Review.
Cited by
-
Sending out an SOS - the bacterial DNA damage response.Genet Mol Biol. 2022 Oct 10;45(3 Suppl 1):e20220107. doi: 10.1590/1678-4685-GMB-2022-0107. eCollection 2022. Genet Mol Biol. 2022. PMID: 36288458 Free PMC article.
-
Interactions and evolutionary relationships among bacterial mobile genetic elements.Nat Rev Microbiol. 2025 Jul;23(7):423-438. doi: 10.1038/s41579-025-01157-y. Epub 2025 Mar 11. Nat Rev Microbiol. 2025. PMID: 40069292 Review.
-
Pathogen-encoded Rum DNA polymerase drives rapid bacterial drug resistance.Nucleic Acids Res. 2024 Nov 27;52(21):12987-13002. doi: 10.1093/nar/gkae899. Nucleic Acids Res. 2024. PMID: 39413207 Free PMC article.
-
Interaction of the SXT/R391 element ICEPmiJpn1 with its natural host Proteus mirabilis.Microbiol Spectr. 2025 Jul;13(7):e0033925. doi: 10.1128/spectrum.00339-25. Epub 2025 May 23. Microbiol Spectr. 2025. PMID: 40407375 Free PMC article.
References
-
- Bie, L. , Wu, H. , Wang, X.H. , Wang, M. & Xu, H. (2017) Identification and characterization of new members of the SXT/R391 family of integrative and conjugative elements (ICEs) in Proteus mirabilis . International Journal of Antimicrobial Agents, 50, 242–246. 10.1016/j.ijantimicag.2017.01.045 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

