KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance
- PMID: 34184733
- PMCID: PMC8181900
- DOI: 10.1242/bio.057729
KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance
Abstract
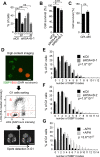
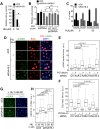
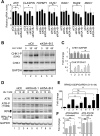
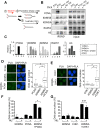
KDM5A and KDM5B histone-demethylases are overexpressed in many cancers and have been involved in drug tolerance. Here, we describe that KDM5A, together with KDM5B, contribute to replication stress (RS) response and tolerance. First, they positively regulate RRM2, the regulatory subunit of ribonucleotide reductase. Second, they are required for optimal levels of activated Chk1, a major player of the intra-S phase checkpoint that protects cells from RS. We also found that KDM5A is enriched at ongoing replication forks and associates with both PCNA and Chk1. Because RRM2 is a major determinant of replication stress tolerance, we developed cells resistant to HU, and show that KDM5A/B proteins are required for both RRM2 overexpression and tolerance to HU. Altogether, our results indicate that KDM5A/B are major players of RS management. They also show that drugs targeting the enzymatic activity of KDM5 proteins may not affect all cancer-related consequences of KDM5A/B overexpression.
Keywords: Chromatin; Drug tolerance; Replication stress.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
-
- Beshiri, M. L., Holmes, K. B., Richter, W. F., Hess, S., Islam, A. B. M. M. K., Yan, Q., Plante, L., Litovchick, L., Gévry, N., Lopez-Bigas, N.et al. (2012). Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc. Natl. Acad. Sci. USA 109, 18499-18504. 10.1073/pnas.1216724109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

