Large-scale sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management
- PMID: 34184982
- PMCID: PMC8461472
- DOI: 10.1099/mgen.0.000589
Large-scale sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management
Abstract
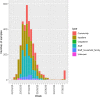
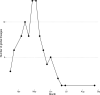
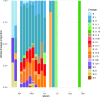
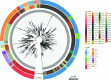
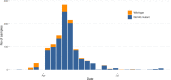
The COVID-19 pandemic has spread rapidly throughout the world. In the UK, the initial peak was in April 2020; in the county of Norfolk (UK) and surrounding areas, which has a stable, low-density population, over 3200 cases were reported between March and August 2020. As part of the activities of the national COVID-19 Genomics Consortium (COG-UK) we undertook whole genome sequencing of the SARS-CoV-2 genomes present in positive clinical samples from the Norfolk region. These samples were collected by four major hospitals, multiple minor hospitals, care facilities and community organizations within Norfolk and surrounding areas. We combined clinical metadata with the sequencing data from regional SARS-CoV-2 genomes to understand the origins, genetic variation, transmission and expansion (spread) of the virus within the region and provide context nationally. Data were fed back into the national effort for pandemic management, whilst simultaneously being used to assist local outbreak analyses. Overall, 1565 positive samples (172 per 100 000 population) from 1376 cases were evaluated; for 140 cases between two and six samples were available providing longitudinal data. This represented 42.6 % of all positive samples identified by hospital testing in the region and encompassed those with clinical need, and health and care workers and their families. In total, 1035 cases had genome sequences of sufficient quality to provide phylogenetic lineages. These genomes belonged to 26 distinct global lineages, indicating that there were multiple separate introductions into the region. Furthermore, 100 genetically distinct UK lineages were detected demonstrating local evolution, at a rate of ~2 SNPs per month, and multiple co-occurring lineages as the pandemic progressed. Our analysis: identified a discrete sublineage associated with six care facilities; found no evidence of reinfection in longitudinal samples; ruled out a nosocomial outbreak; identified 16 lineages in key workers which were not in patients, indicating infection control measures were effective; and found the D614G spike protein mutation which is linked to increased transmissibility dominates the samples and rapidly confirmed relatedness of cases in an outbreak at a food processing facility. The large-scale genome sequencing of SARS-CoV-2-positive samples has provided valuable additional data for public health epidemiology in the Norfolk region, and will continue to help identify and untangle hidden transmission chains as the pandemic evolves.
Keywords: ARTIC; NGS; SARS-CoV-2; genome; genomic epidemiology; sequencing.
Conflict of interest statement
L.G. received a partial support for his PhD from Roche. The other authors declare that there are no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

