Genomewide Association Studies in Pharmacogenomics
- PMID: 34185318
- PMCID: PMC8376796
- DOI: 10.1002/cpt.2349
Genomewide Association Studies in Pharmacogenomics
Abstract
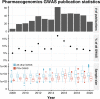
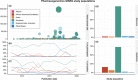
The increasing availability of genotype data linked with information about drug-response phenotypes has enabled genomewide association studies (GWAS) that uncover genetic determinants of drug response. GWAS have discovered associations between genetic variants and both drug efficacy and adverse drug reactions. Despite these successes, the design of GWAS in pharmacogenomics (PGx) faces unique challenges. In this review, we analyze the last decade of GWAS in PGx. We review trends in publications over time, including the drugs and drug classes studied and the clinical phenotypes used. Several data sharing consortia have contributed substantially to the PGx GWAS literature. We anticipate increased focus on biobanks and highlight phenotypes that would best enable future PGx discoveries.
© 2021 The Authors. Clinical Pharmacology & Therapeutics published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
R.B.A. is a stockholder in Personalis.com, 23andMe.com. All other authors declared no competing interests for this work.
Figures