Comparative Host Interactomes of the SARS-CoV-2 Nonstructural Protein 3 and Human Coronavirus Homologs
- PMID: 34186245
- PMCID: PMC8236078
- DOI: 10.1016/j.mcpro.2021.100120
Comparative Host Interactomes of the SARS-CoV-2 Nonstructural Protein 3 and Human Coronavirus Homologs
Abstract
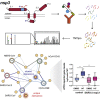
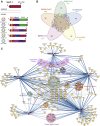
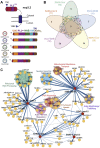
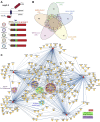
Human coronaviruses have become an increasing threat to global health; three highly pathogenic strains have emerged since the early 2000s, including most recently SARS-CoV-2, the cause of COVID-19. A better understanding of the molecular mechanisms of coronavirus pathogenesis is needed, including how these highly virulent strains differ from those that cause milder, common-cold-like disease. While significant progress has been made in understanding how SARS-CoV-2 proteins interact with the host cell, nonstructural protein 3 (nsp3) has largely been omitted from the analyses. Nsp3 is a viral protease with important roles in viral protein biogenesis, replication complex formation, and modulation of host ubiquitinylation and ISGylation. Herein, we use affinity purification-mass spectrometry to study the host-viral protein-protein interactome of nsp3 from five coronavirus strains: pathogenic strains SARS-CoV-2, SARS-CoV, and MERS-CoV; and endemic common-cold strains hCoV-229E and hCoV-OC43. We divide each nsp3 into three fragments and use tandem mass tag technology to directly compare the interactors across the five strains for each fragment. We find that few interactors are common across all variants for a particular fragment, but we identify shared patterns between select variants, such as ribosomal proteins enriched in the N-terminal fragment (nsp3.1) data set for SARS-CoV-2 and SARS-CoV. We also identify unique biological processes enriched for individual homologs, for instance, nuclear protein import for the middle fragment of hCoV-229E, as well as ribosome biogenesis of the MERS nsp3.2 homolog. Lastly, we further investigate the interaction of the SARS-CoV-2 nsp3 N-terminal fragment with ATF6, a regulator of the unfolded protein response. We show that SARS-CoV-2 nsp3.1 directly binds to ATF6 and can suppress the ATF6 stress response. Characterizing the host interactions of nsp3 widens our understanding of how coronaviruses co-opt cellular pathways and presents new avenues for host-targeted antiviral therapeutics.
Keywords: COVID-19; activating transcription factor 6; affinity purification-mass spectrometry; nsp3; tandem mass tags; unfolded protein response.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures

Update of
-
Comparative host interactomes of the SARS-CoV-2 nonstructural protein 3 and human coronavirus homologs.bioRxiv [Preprint]. 2021 Mar 8:2021.03.08.434440. doi: 10.1101/2021.03.08.434440. bioRxiv. 2021. Update in: Mol Cell Proteomics. 2021;20:100120. doi: 10.1016/j.mcpro.2021.100120. PMID: 33758849 Free PMC article. Updated. Preprint.
References
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

