Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis
- PMID: 34187846
- PMCID: PMC9120377
- DOI: 10.1136/gutjnl-2020-323906
Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis
Abstract
Objective: Gastro-oesophageal reflux disease (GERD) has heterogeneous aetiology primarily attributable to its symptom-based definitions. GERD genome-wide association studies (GWASs) have shown strong genetic overlaps with established risk factors such as obesity and depression. We hypothesised that the shared genetic architecture between GERD and these risk factors can be leveraged to (1) identify new GERD and Barrett's oesophagus (BE) risk loci and (2) explore potentially heterogeneous pathways leading to GERD and oesophageal complications.
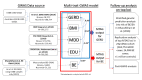
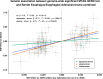
Design: We applied multitrait GWAS models combining GERD (78 707 cases; 288 734 controls) and genetically correlated traits including education attainment, depression and body mass index. We also used multitrait analysis to identify BE risk loci. Top hits were replicated in 23andMe (462 753 GERD cases, 24 099 BE cases, 1 484 025 controls). We additionally dissected the GERD loci into obesity-driven and depression-driven subgroups. These subgroups were investigated to determine how they relate to tissue-specific gene expression and to risk of serious oesophageal disease (BE and/or oesophageal adenocarcinoma, EA).
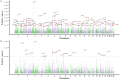
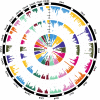
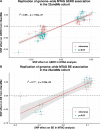
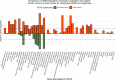
Results: We identified 88 loci associated with GERD, with 59 replicating in 23andMe after multiple testing corrections. Our BE analysis identified seven novel loci. Additionally we showed that only the obesity-driven GERD loci (but not the depression-driven loci) were associated with genes enriched in oesophageal tissues and successfully predicted BE/EA.
Conclusion: Our multitrait model identified many novel risk loci for GERD and BE. We present strong evidence for a genetic underpinning of disease heterogeneity in GERD and show that GERD loci associated with depressive symptoms are not strong predictors of BE/EA relative to obesity-driven GERD loci.
Keywords: Barrett's oesophagus; gastro-esophageal reflux disease; genetics; oesophageal reflux.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: Authors listed in the 23andMe Research Team are employees for the company 23andMe Co.
Figures
Comment in
-
Causal relationship between air pollution, lung function, gastroesophageal reflux disease, and non-alcoholic fatty liver disease: univariate and multivariate Mendelian randomization study.Front Public Health. 2024 Apr 29;12:1368483. doi: 10.3389/fpubh.2024.1368483. eCollection 2024. Front Public Health. 2024. PMID: 38746002 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
