Chromosome anchoring in Senegalese sole (Solea senegalensis) reveals sex-associated markers and genome rearrangements in flatfish
- PMID: 34188074
- PMCID: PMC8242048
- DOI: 10.1038/s41598-021-92601-5
Chromosome anchoring in Senegalese sole (Solea senegalensis) reveals sex-associated markers and genome rearrangements in flatfish
Abstract
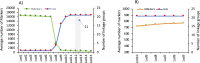
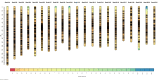
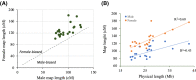
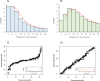
The integration of physical and high-density genetic maps is a very useful approach to achieve chromosome-level genome assemblies. Here, the genome of a male Senegalese sole (Solea senegalensis) was de novo assembled and the contigs were anchored to a high-quality genetic map for chromosome-level scaffolding. Hybrid assembled genome was 609.3 Mb long and contained 3403 contigs with a N50 of 513 kb. The linkage map was constructed using 16,287 informative SNPs derived from ddRAD sequencing in 327 sole individuals from five families. Markers were assigned to 21 linkage groups with an average number of 21.9 markers per megabase. The anchoring of the physical to the genetic map positioned 1563 contigs into 21 pseudo-chromosomes covering 548.6 Mb. Comparison of genetic and physical distances indicated that the average genome-wide recombination rate was 0.23 cM/Mb and the female-to-male ratio 1.49 (female map length: 2,698.4 cM, male: 2,036.6 cM). Genomic recombination landscapes were different between sexes with crossovers mainly concentrated toward the telomeres in males while they were more uniformly distributed in females. A GWAS analysis using seven families identified 30 significant sex-associated SNP markers located in linkage group 18. The follicle-stimulating hormone receptor appeared as the most promising locus associated with sex within a region with very low recombination rates. An incomplete penetrance of sex markers with males as the heterogametic sex was determined. An interspecific comparison with other Pleuronectiformes genomes identified a high sequence similarity between homologous chromosomes, and several chromosomal rearrangements including a lineage-specific Robertsonian fusion in S. senegalensis.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Liu ZJ, Cordes FJ. DNA marker technology and their applications in aquaculture genetics. Aquaculture. 2004;238:1–37. doi: 10.1016/j.aquaculture.2004.05.027. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

