Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding
- PMID: 34191293
- PMCID: PMC8327961
- DOI: 10.15252/embj.2021107807
Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding
Abstract
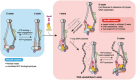
Eukaryotic cells employ three SMC (structural maintenance of chromosomes) complexes to control DNA folding and topology. The Smc5/6 complex plays roles in DNA repair and in preventing the accumulation of deleterious DNA junctions. To elucidate how specific features of Smc5/6 govern these functions, we reconstituted the yeast holo-complex. We found that the Nse5/6 sub-complex strongly inhibited the Smc5/6 ATPase by preventing productive ATP binding. This inhibition was relieved by plasmid DNA binding but not by short linear DNA, while opposing effects were observed without Nse5/6. We uncovered two binding sites for Nse5/6 on Smc5/6, based on an Nse5/6 crystal structure and cross-linking mass spectrometry data. One binding site is located at the Smc5/6 arms and one at the heads, the latter likely exerting inhibitory effects on ATP hydrolysis. Cysteine cross-linking demonstrated that the interaction with Nse5/6 anchored the ATPase domains in a non-productive state, which was destabilized by ATP and DNA. Under similar conditions, the Nse4/3/1 module detached from the ATPase. Altogether, we show how DNA substrate selection is modulated by direct inhibition of the Smc5/6 ATPase by Nse5/6.
Keywords: Smc5/6; chromosome segregation; cohesion; condensin; loop extrusion.
© 2021 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
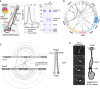
Reconstitution of Smc5/6 and Nse5/6 complexes. Left panel, schematic depiction of the composition and organization of the yeast Smc5/6 “core” hexamer and the Nse5/6 dimer. Middle panel, elution profiles for analytical gel filtration (Superose 6 Increase 3.2/300) of the Smc5/6 core hexamer, the Nse5/6 dimer and a holo‐complex obtained by mixing of dimer and hexamer. Measured by absorption at 280 nm. Right panel, peak fractions were analysed by SDS–PAGE and Coomassie Brilliant Blue staining.
Circular representation of lysine–lysine inter‐subunit cross‐links identified by mass spectrometry (XL‐MS) in buffer containing 250 mM NaCl. For simplicity, cross‐links between proteins (or domains) are grouped, and the thickness of the lines indicates the total number of cross‐links of this particular type. Smc5 and Smc6 proteins are divided into N‐ and C‐terminal head domains (HD) and coiled‐coil arms (CC) as well as the central hinge domain. For a full representation of individual inter‐subunit as well as intra‐subunit cross‐links, see Dataset EV1.
Cross‐links obtained within and between Smc5 and Smc6 subunits of the Smc5/6 octamer. A cross‐link that did not match to the elongated rod‐shaped particle is displayed by a dashed line in red colours. The cartoon on the right shows the dimer of folded Smc5 and Smc6 proteins, with examples of intra‐ and inter‐links within the coiled‐coil arms indicated with white and black lines, respectively.
Selected 2D class averages obtained by cryo‐electron microscopy of the yeast Smc5/6 hexamer (left images). Representative classes with dimers of Smc5/6 hexamers are displayed (images 3 and 4). Emerging details are indicated schematically (right panel).

Domain organization of Nse5 and Nse6 proteins. Putative domain boundaries identified by secondary structure prediction and limited proteolysis are denoted by arrowheads, in black colours when producing stable fragments, otherwise in grey colours.
Limited proteolysis of a purified preparation of the Nse5/Nse6 complex by trypsin. Samples taken at different time points were analysed by SDS–PAGE and Coomassie staining. Selected stable fragments identified by mass spectrometry are indicated.
Front and back view of the Nse5/Nse6 co‐crystal structure in cartoon representation. Structural elements of Nse5 and Nse6 are displayed in orange and purple colours, respectively. α‐helices are labelled.
Conservation of residues at the Nse5/6 binding interface. Interaction surface of Nse6 (left panel) and Nse5 (right panel). For orientation, the secondary structure at the interface is displayed (middle panel). Colour code for residue conservation is given at the bottom of the panel.
A conserved surface area (bottom panel) on top of Nse5/6. Colour coding for conserved residues as in (D). An extended loop in this region contains a lysine residue (K148) that was found to cross‐link to Smc5 and Smc6 ATPase heads (see Fig 3).

The Nse6 moiety of the Nse5/6 dimer structure is shown in front, back and side views in cartoon representation and in corresponding surface conservation displays (at the bottom left of each panel). The concave surface interacting with Nse5 shows highest residue conservation. Conservation colour code is indicated at the right.
Front and back views of Nse5 in the Nse5/6 structure in cartoon and corresponding surface conservation representation. Display as in (A). Conservation colour code as in (A).
Zoom view of contacts at the Nse5/6 interface formed by residues in helix α12 as well as the preceding loop in Nse5 with Nse6 helices α1 and α3.
Verification of the Nse5/6 structure by BMOE cross‐linking. Residues H368 in Nse6 and G56 in Nse5 (see structure with zoom‐in on the left) were mutated to cysteines. Incubation with BMOE (30 s) of the mutant but not the wild‐type complex led to robust cross‐linking seen as a decrease in electrophoretic mobility of the cross‐linked species in SDS–PAGE (right).
(left panel) Superimposition of Nse6 helices α8‐α11 with selected top hits from a DALI search in the Protein Data Bank in top and front view. Root mean square displacement (rmsd) values are given as indicator for the quality of the fit. (right panel) Similar analysis for the N‐terminal region of Nse5 (helices α1‐ α6).
Superimposition of the Nse5/6 crystal structure (this work; PDB: 7OGG) with a similar cryo‐EM structure published while this work was in progress (PDB: 7LTO). The two independently derived models are highly similar (rmsd 0.829 Å).

Competition binding of a mixture of Nse5‐His/Nse6 and Nse6(1‐179)‐CPD‐His (“in”) to immobilized Smc5/Smc6‐Twin‐Strep hexamer. Input (“in”) and pulldown fractions were analysed by SDS–PAGE and Coomassie staining.
Competition with excess Nse5/6 and Nse6 fragment. Smc5/6 hexamers were immobilized together with Nse5‐His/Nse6 or Nse6(1–179)‐CPD‐His and washed with Nse6(1‐179)‐CPD‐His or Nse5‐His/Nse6, respectively. Input (“in”), pulldown and control pulldowns (without competition) were analysed by SDS–PAGE and Coomassie staining.
Pulldown (“pd”) assays examining the interaction of Smc5/Smc6‐Twin‐Strep (“TS”) hexamer (“in”) with either Nse6(1–179)‐CPD‐His or Nse6(86–179)‐CPD‐His. Fractions were analysed by SDS–PAGE and Coomassie staining showing that residues 1–85 in Nse6 are dispensable for interaction with the Smc5/6 hexamer.
Spotting assay to determine sensitivity of yeast strains to UV irradiation and treatment with methyl methanesulphonate (MMS) or hydroxyurea (HU). A strain lacking the N‐terminal region of Nse6 (nse6(86‐C)) does not show increased sensitivity compared to the wild type, while a control strain (smc6(R135E)) recently described by others (Serrano et al, 2020) does.

Pulldown (“pd”) assays using immobilized Smc5/Smc6‐Twin‐Strep (“TS”) hexamers and soluble input material (“in”) of Nse5/6 (left), of Nse5/Nse6(177‐C) (middle) and of Nse6(1‐179)‐CPD‐His (right). Control pulldowns (“−”) were performed by omitting the pre‐binding of Smc5/Smc6‐Twin‐Strep to the beads. Fractions were analysed by SDS–PAGE and Coomassie staining.
Salt stability of Smc5/Smc6‐Twin‐Strep interactions with Nse5/6 and Nse6(1–179). Immobilized Smc5/Smc6‐Twin‐Strep was mixed with Nse5/6 or Nse6(1–179)‐CPD‐His (“in”). Beads were washed with buffers containing the indicated salt concentrations. Bead fractions were analysed by SDS–PAGE and Coomassie staining. Pulldown efficiencies were estimated from the intensity of Coomassie gel bands.
XL‐MS cross‐links detected between Nse5 and Nse6 proteins and subunits of the Smc5/6 hexamer in schematic representation (middle panel). The positions of lysine residues on the Smc6 head and the Smc5 head, left and right panels, respectively, with cross‐links to Nse5 are denoted on Phyre2‐generated homology models.
Analysis of spore viability by yeast tetrad dissection. Diploid strains heterozygous for alleles of Nse6 (wt, 86‐C, 177‐C and 179‐C) were sporulated. Isolated spores were grown on YPD plates. Viable clones were tested for the marker cassette conferring resistance to G418 (marked by circles in green colours). Dead spores were marked by circles in red colours.

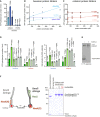
ATP hydrolysis rates (given per Smc5/6 complex) were measured by an enzyme‐coupled assay in the absence and presence of plasmidDNA (conc. 1.25 nM) for the Smc5/6 hexamer (conc. 150 nM) and the octamer (conc. 150 nM) with increasing concentrations of ATP. The curves were fitted to Michaelis–Menten equation, and Km and kcat values were determined. Please note that the hexamer without plasmidDNA shows cooperative behaviour; thus, the Michaelis–Menten kinetics is not formally applicable (marked by asterisk). Assays were performed in biological triplicates, and mean values are shown with error bars indicating standard deviations. For bar graphs, individual data points are also displayed.
Same as in (A) using 40bpDNA (annealed 40 bp oligo DNA) (conc. 1 µM) instead of plasmidDNA. Note that the data points and curves for samples without DNA (hexamer ‐DNA; octamer ‐DNA) are identical to (A).
Fluorescence anisotropy measurements using 40 bp dsDNA or 40‐mer ssDNA substrates. Representative binding curves (top graph) and the resulting Kd values (bottom graph) indicate that both hexameric and octameric Smc5/6 complexes interact with both substrates, whereas the Nse5/6 complex alone does not. The bar graph shows mean values with standard deviations from technical triplicates. Individual data points are also displayed.
Pulldown experiments with Smc5/6 complexes and circular plasmid (2.8 kbp). DNA is retained after high salt washes (1 M NaCl) only in the presence of both Nse5/6 and ATP. The graph on the right shows a quantification (mean values and standard deviations) of the amount of co‐purified DNA from technical triplicates. Individual data points are also displayed.
Salt‐stable DNA association requires the DNA substrate to be circular (“circ”). Same experimental setup as in (D), but the plasmid substrate was also linearized by restriction digest (“lin”).
Salt‐stable DNA association requires ATP binding and Smc5/6 head engagement, but not ATP hydrolysis. Same experimental setup as in (D) using mutant versions of the Smc5/6 hexamer as well as the non‐hydrolysable analogue of ATP, ATPγS, as indicated.
ATP hydrolysis rates for wild‐type Smc5/6 hexamers and the Smc5(EQ)/Smc6(EQ) variant. Rates for the absence and presence of DNA substrates are given (per Smc complex per minute). Assays were performed in technical triplicates, and mean values are shown with error bars indicating standard deviations. Individual data points are also displayed.
Cooperativity in ATP hydrolysis by the Smc5/6 hexamer. ATP hydrolysis rates were determined at different protein concentrations. Smc5/6 hexamers showed cooperative behaviour without DNA and with 40bpDNA but not with plasmidDNA. Assays were performed in technical triplicates, and individual data points are displayed.
Absence of cooperative in ATP hydrolysis by the Smc5/6 octamer. As in (B) using reconstituted octamers. Assays were performed in technical triplicates, and individual data points are displayed.
Overview of values for ATP hydrolysis rates measured at an elevated ATP concentration (1 mM) (left panel) and fold‐stimulation of ATP hydrolysis rates by addition of 40bpDNA (conc. 1 µM), plasmidDNA (conc. 1.5 nM) in closed covalent circular (“circ”) or linearized form (“lin”) (right panel), or 40merssDNA (conc. 1 µM). Assays were performed in technical triplicates, and mean values are shown with error bars indicating standard deviations. Individual data points are also displayed.
Analysis of the plasmid used in ATPase assays on a 0.7% agarose gel stained with SybrSafe, either untreated or after treatment with Topoisomerase I. The plasmid is present as a mixture of supercoiled and relaxed form. Note that due to the large size the relaxed form does not properly enter the gel and is largely retained in the wells.
Pulldown assay between hinge‐less Smc5 and Smc6 sub‐complexes carrying the “EQ” mutation that abolishes ATP hydrolysis (Smc5Δhinge(EQ)/Nse2/Nse4(C) and Smc6Δhinge(EQ)/Nse4N), respectively; see scheme on the left) in the presence or absence of ATP or ATPγS. The SDS–PAGE gel on the right shows that the complexes do not detectably interact in the absence of ATP but do so in its presence. Head engagement is clearly less efficient with the non‐hydrolysable analogue ATPγS.

Cross‐linking of a purified preparation of wild‐type Smc5/6 hexamer in the absence and presence of Nse5/6. ATP and plasmidDNA were added as substrates as denoted. Products were analysed by SDS–PAGE and Coomassie staining. A cross‐linked species derived from natural cysteines is indicated by an arrowhead (“XL”). Note that a fraction of Nse4 is converted into a more slowly migrating species, presumably by intra‐molecular cross‐linking.
Cross‐linking of engineered variants of the Smc5/6 hexamer. As in (A) using Smc5 and Smc6 cysteine mutants. Schemes indicate the location of engineered cysteines and their expected ability to cross‐link in a rod‐like and a ring‐like conformation. High‐molecular weight species were analysed by SDS–PAGE and Coomassie staining. Wild‐type hexamer (“wt”) is included as cross‐linking control. Species occurring only in the presence of engineered cysteines are labelled by coloured arrowheads. Cross‐linking efficiencies were calculated from the intensity of Coomassie‐stained bands by comparing the band of the corresponding cross‐linked species to the bands of unmodified Smc5 and Smc6. Numbers below the gel quantify the percentage of cross‐linked protein species in the displayed gel. Comparable numbers were obtained in at least one additional independent experiment.
DNA effects on Smc5/6 arm co‐alignment as judged by CC‐Cys cross‐linking. As in (B) but also including 40bpDNA. Addition of plasmidDNA but not 40bpDNA reduces CC‐Cys cross‐linking in the Smc5/6 hexamer.
DNA effects on head engagement as judged by E‐Cys cross‐linking. As in (B) also including 40bpDNA. plasmidDNA binding, but not 40bpDNA binding, overcomes inhibition of E‐Cys cross‐linking by Nse5/6.
Effects of 40bpDNA and plasmidDNA on head engagement (as judged by E‐Cys cross‐linking) of the Smc5/6 complex with wild type (wt) or hydrolysis‐deficient (EQ) heads, in both the presence or the absence of the Nse5/6 complex. Cross‐linking efficiencies were calculated from the intensity of Coomassie‐stained bands by comparing the band of the corresponding cross‐linked species to the bands of unmodified Smc5 and Smc6. Numbers below the gel quantify the percentage of cross‐linked protein species in the displayed gel.
Effects of plasmidDNA on head engagement of the Smc5/6 complex (as judged by E‐Cys cross‐linking) in the presence of ATP or the non‐hydrolysable analogue ATPγS. Calculations and quantifications as in (E).

Hinge‐Cys residues were chosen based on a homology model of the budding yeast Smc5/6 hinge domain built from the fission yeast hinge structure (Alt et al, 2017). Residues for cross‐linking of the “south” interface (top; Smc5(N526C) and Smc6(N643C)) or the “north” interface (bottom; Smc5(V638C) and Smc6(N572C)) are indicated in green colours on the hinge structure in surface representation.
Hinge‐Cys cross‐linking at the “north” hinge interface. Scheme indicates the location of engineered cysteines and their expected ability to cross‐link in a rod‐like and a ring‐like conformation. High‐molecular weight species were analysed by SDS–PAGE and Coomassie staining. Wild‐type hexamer (“wt”) is included as cross‐linking control. Species occurring only in the presence of engineered cysteines are labelled by coloured arrowheads. Cross‐linking efficiencies were calculated from the intensity of Coomassie‐stained bands by comparing the band of the corresponding cross‐linked species to the bands of unmodified Smc5 and Smc6. Numbers below the gel quantify the percentage of cross‐linked protein species in the displayed gel.
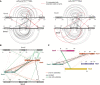
Strategy for the construction of models for the J‐state and the E‐state based on structural models of the archaeal Smc‐ScpAB complex.
Positions of J‐Cys (in blue colours), CC‐Cys (in purple colours) and E‐Cys (in red colours) residues on models of the J‐state and the E‐state.
E‐Cys cross‐linking in the context of an ATP hydrolysis‐deficient (EQ) Smc5/6 complex. Rest as in EV4B.
Cross‐links in the Smc5/6 octamer identified by XL‐MS without substrate addition (left panel) and with plasmidDNA and ATP (right panel). As in Fig 1C using ATPase buffer. Only intra‐ and inter‐links of the Smc5/6 dimer are displayed here. Other cross‐links are shown in panel (B), (C), and Fig EV5 and listed in Dataset EV1.
Cross‐links of Nse3/4 detected by XL‐MS. Same experiment as in (A) using the Smc5/6 octamer in ATPase buffer. Cross‐links between Nse3 and Nse4 as well as their cross‐links to Smc5 and Smc6 are displayed. Lines in black colours indicate cross‐links observed with and without substrates, in oranges colours only without substrates and in dashed lines in green colours only with substrate addition. See Fig EV5B for an alternative representation in circular plots.
Cross‐links of Nse1/3/4 to Nse5/6. Same experiment as in (A) and (B). Display as in (B). See Fig EV5B for an alternative representation in circular plots.

Cross‐links between Nse2 and Smc5/Smc6 proteins remain unaltered upon plasmidDNA addition.
Changes in inter‐subunit cross‐links between Nse1/3/4 (left) and Nse5/6 (right) modules. Alternative representation of the same data shown in Fig 6B and C.
Cross‐links between Nse3 and Nse4 and between Nse3/Nse4 and Smc5/Smc6 detected in the Smc5(EQ)/Smc6(EQ) octamer without ATP and plasmidDNA (left panel) and with ATP and plasmidDNA (right panel).
Same as in (C) for cross‐links between Nse1/3/4/5/6 proteins.
Pulldowns of the octameric complex (“in”) via Smc6‐Twin‐Strep in the absence (“−”) or presence (“+”) of ATP and plasmid DNA under the same conditions as the ones used for XL‐MS analysis. The Nse4/3/1 module is still present in the complex despite the severely reduced cross‐linking efficiency.
References
-
- Adamus M, Lelkes E, Potesil D, Ganji SR, Kolesar P, Zabrady K, Zdrahal Z, Palecek JJ (2020) Molecular Insights into the Architecture of the Human SMC5/6 Complex. J Mol Biol 432: 3820–3837 - PubMed
-
- Aragon L (2018) The Smc5/6 complex: new and old functions of the enigmatic long‐distance relative. Annu Rev Genet 52: 89–107 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

