Cryptic association of B7-2 molecules and its implication for clustering
- PMID: 34191384
- PMCID: PMC8376414
- DOI: 10.1002/pro.4151
Cryptic association of B7-2 molecules and its implication for clustering
Abstract
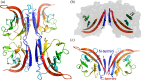
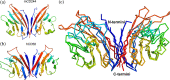
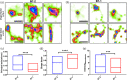
T-cell co-stimulation through CD28/CTLA4:B7-1/B7-2 axis is one of the extensively studied pathways that resulted in the discovery of several FDA-approved drugs for autoimmunity and cancer. However, many aspects of the signaling mechanism remain elusive, including oligomeric association and clustering of B7-2 on the cell surface. Here, we describe the structure of the IgV domain of B7-2 and its cryptic association into 1D arrays that appear to represent the pre-signaling state of B7-2 on the cell membrane. Super-resolution microscopy experiments on heterologous cells expressing B7-2 and B7-1 suggest, B7-2 form relatively elongated and larger clusters compared to B7-1. The sequence and structural comparison of other B7 family members, B7-1:CTLA4 and B7-2:CTLA-4 complex structures, support our view that the observed B7-2 1D zipper array is physiologically important. This observed 1D zipper-like array also provides an explanation for its clustering, and upright orientation on the cell surface, and avoidance of spurious signaling.
Keywords: B7-2 structure; immune checkpoint blockade; immune receptors; protein clustering.
© 2021 The Protein Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
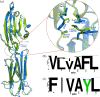


References
-
- Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. - PubMed
-
- Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975;53:27–42. - PubMed
-
- Babcock SK, Gill RG, Bellgrau D, Lafferty KJ. Studies on the two‐signal model for T cell activation in vivo. Transplant Proc. 1987;19:303–306. - PubMed
-
- Carreno BM, Collins M. The B7 family of ligands and its receptors: New pathways for costimulation and inhibition of immune responses. Annu Rev Immunol. 2002;20:29–53. - PubMed
-
- Grakoui A, Bromley SK, Sumen C, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285:221–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

