Role of Gut Microbiota in Multiple Sclerosis and Potential Therapeutic Implications
- PMID: 34191698
- PMCID: PMC9881072
- DOI: 10.2174/1570159X19666210629145351
Role of Gut Microbiota in Multiple Sclerosis and Potential Therapeutic Implications
Abstract
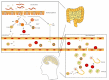
The role of gut microbiota in health and diseases has been receiving increased attention recently. Emerging evidence from previous studies on gut-microbiota-brain axis highlighted the importance of gut microbiota in neurological disorders. Multiple sclerosis (MS) is a chronic, inflammatory, demyelinating disease of the central nervous system (CNS) resulting from T-cell-driven, myelin-directed autoimmunity. The dysbiosis of gut microbiota in MS patients has been reported in published research studies, indicating that gut microbiota plays an important role in the pathogenesis of MS. Gut microbiota have also been reported to influence the initiation of disease and severity of experimental autoimmune encephalomyelitis, which is the animal model of MS. However, the underlying mechanisms of gut microbiota involvement in the pathogenesis of MS remain unclear. Therefore, in this review, we summerized the potential mechanisms for gut microbiota involvement in the pathogenesis of MS, including increasing the permeability of the intestinal barrier, initiating an autoimmune response, disrupting the blood-brain barrier integrity, and contributing to chronic inflammation. The possibility for gut microbiota as a target for MS therapy has also been discussed. This review provides new insight into understanding the role of gut microbiota in neurological and inflammatory diseases.
Keywords: Gut microbiota; antibiotic treatment; blood-brain barrier; fecal microbiota transplantation; gut-microbiota-brain axis; multiple sclerosis; neuro-inflammatory diseases; probiotic microbiota.
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Figures
References
-
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.M., Hansen T., Le Pasli-er D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissen-bach J., Bork P., Ehrlich S.D., Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

