Light-induced changes in the suprachiasmatic nucleus transcriptome regulated by the ERK/MAPK pathway
- PMID: 34191798
- PMCID: PMC8244859
- DOI: 10.1371/journal.pone.0249430
Light-induced changes in the suprachiasmatic nucleus transcriptome regulated by the ERK/MAPK pathway
Abstract
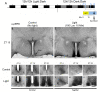
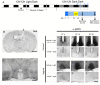
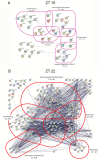
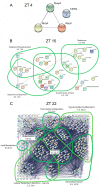
The mammalian master circadian pacemaker within the suprachiasmatic nucleus (SCN) maintains tight entrainment to the 24 hr light/dark cycle via a sophisticated clock-gated rhythm in the responsiveness of the oscillator to light. A central event in this light entrainment process appears to be the rapid induction of gene expression via the ERK/MAPK pathway. Here, we used RNA array-based profiling in combination with pharmacological disruption methods to examine the contribution of ERK/MAPK signaling to light-evoked gene expression. Transient photic stimulation during the circadian night, but not during the circadian day, triggered marked changes in gene expression, with early-night light predominately leading to increased gene expression and late-night light predominately leading to gene downregulation. Functional analysis revealed that light-regulated genes are involved in a diversity of physiological processes, including DNA transcription, RNA translation, mRNA processing, synaptic plasticity and circadian timing. The disruption of MAPK signaling led to a marked reduction in light-evoked gene regulation during the early night (32/52 genes) and late night (190/191 genes); further, MAPK signaling was found to gate gene expression across the circadian cycle. Together, these experiments reveal potentially important insights into the transcriptional-based mechanisms by which the ERK/MAPK pathway regulates circadian clock timing and light-evoked clock entrainment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

