SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2
- PMID: 34192529
- PMCID: PMC8185186
- DOI: 10.1016/j.cell.2021.06.005
SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2
Abstract
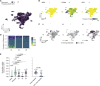
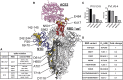
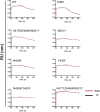
In this study we profiled vaccine-induced polyclonal antibodies as well as plasmablast-derived mAbs from individuals who received SARS-CoV-2 spike mRNA vaccine. Polyclonal antibody responses in vaccinees were robust and comparable to or exceeded those seen after natural infection. However, the ratio of binding to neutralizing antibodies after vaccination was greater than that after natural infection and, at the monoclonal level, we found that the majority of vaccine-induced antibodies did not have neutralizing activity. We also found a co-dominance of mAbs targeting the NTD and RBD of SARS-CoV-2 spike and an original antigenic-sin like backboost to spikes of seasonal human coronaviruses OC43 and HKU1. Neutralizing activity of NTD mAbs but not RBD mAbs against a clinical viral isolate carrying E484K as well as extensive changes in the NTD was abolished, suggesting that a proportion of vaccine-induced RBD binding antibodies may provide substantial protection against viral variants carrying single E484K RBD mutations.
Keywords: NTD; RBD; SARS-CoV-2; mAbs; mRNA vaccination; plasmablasts; spike.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list F.K. as co-inventor. V.S. and F.A. are also listed on the serological assay patent application as a co-inventors. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. F.K. has consulted for Merck and Pfizer (before 2020) and is currently consulting for Pfizer, Seqirus, and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2. A.H.E. has consulted for InBios and Fimbrion Therapeutics (before 2021) and is currently a consultant for Mubadala Investment Company. The Ellebedy laboratory received funding under sponsored research agreements that are unrelated to the data presented in the current study from Emergent BioSolutions and from AbbVie.
Figures







References
-
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 2021 doi: 10.1101/2021.02.23.21252259. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

