Surprising Twists in Nucleosomal DNA with Implication for Higher-order Folding
- PMID: 34192585
- PMCID: PMC8380738
- DOI: 10.1016/j.jmb.2021.167121
Surprising Twists in Nucleosomal DNA with Implication for Higher-order Folding
Abstract
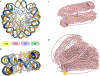
While nucleosomes are dynamic entities that must undergo structural deformations to perform their functions, the general view from available high-resolution structures is a largely static one. Even though numerous examples of twist defects have been documented, the DNA wrapped around the histone core is generally thought to be overtwisted. Analysis of available high-resolution structures from the Protein Data Bank reveals a heterogeneous distribution of twist along the nucleosomal DNA, with clear patterns that are consistent with the literature, and a significant fraction of structures that are undertwisted. The subtle differences in nucleosomal DNA folding, which extend beyond twist, have implications for nucleosome disassembly and modeled higher-order structures. Simulations of oligonucleosome arrays built with undertwisted models behave very differently from those constructed from overtwisted models, in terms of compaction and inter-nucleosome contacts, introducing configurational changes equivalent to those associated with 2-3 base-pair changes in nucleosome spacing. Differences in the nucleosomal DNA pathway, which underlie the way that DNA enters and exits the nucleosome, give rise to different nucleosome-decorated minicircles and affect the topological mix of configurational states.
Keywords: DNA minicircle; Monte Carlo DNA simulation; nucleosomal twist uptake; nucleosome gaping; oligonucleosome array; undertwisted nucleosome.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest Statement The authors declare no conflicts of interest.
Figures

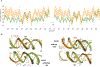




References
-
- Polach KJ & Widom J (1995). Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol 254, 130–149. - PubMed
-
- Mohapatra S, Lin C-T, Feng XA, Basu A & Ha T (2020). Single-molecule analysis and engineering of DNA motors. Chem. Rev 120, 36–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

