Probing the SAM Binding Site of SARS-CoV-2 Nsp14 In Vitro Using SAM Competitive Inhibitors Guides Developing Selective Bisubstrate Inhibitors
- PMID: 34192965
- PMCID: PMC8458670
- DOI: 10.1177/24725552211026261
Probing the SAM Binding Site of SARS-CoV-2 Nsp14 In Vitro Using SAM Competitive Inhibitors Guides Developing Selective Bisubstrate Inhibitors
Abstract
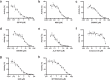
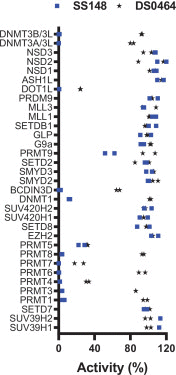
The COVID-19 pandemic has clearly brought the healthcare systems worldwide to a breaking point, along with devastating socioeconomic consequences. The SARS-CoV-2 virus, which causes the disease, uses RNA capping to evade the human immune system. Nonstructural protein (nsp) 14 is one of the 16 nsps in SARS-CoV-2 and catalyzes the methylation of the viral RNA at N7-guanosine in the cap formation process. To discover small-molecule inhibitors of nsp14 methyltransferase (MTase) activity, we developed and employed a radiometric MTase assay to screen a library of 161 in-house synthesized S-adenosylmethionine (SAM) competitive MTase inhibitors and SAM analogs. Among six identified screening hits, SS148 inhibited nsp14 MTase activity with an IC50 value of 70 ± 6 nM and was selective against 20 human protein lysine MTases, indicating significant differences in SAM binding sites. Interestingly, DS0464 with an IC50 value of 1.1 ± 0.2 µM showed a bisubstrate competitive inhibitor mechanism of action. DS0464 was also selective against 28 out of 33 RNA, DNA, and protein MTases. The structure-activity relationship provided by these compounds should guide the optimization of selective bisubstrate nsp14 inhibitors and may provide a path toward a novel class of antivirals against COVID-19, and possibly other coronaviruses.
Keywords: COVID-19; SARS-CoV-2; coronavirus; nsp14.
Figures
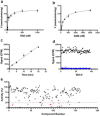
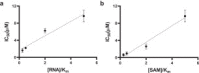
 ) and presence (
) and presence ( ) of 1 µM sinefungin. (e) Nsp14 was screened against a library of SAM analogs. A collection of 161 SAM competitive inhibitors and analogs was tested at 50 µM for inhibition of nsp14 activity under conditions described for Z′-factor determination. Compounds with higher than 75% inhibition (
) of 1 µM sinefungin. (e) Nsp14 was screened against a library of SAM analogs. A collection of 161 SAM competitive inhibitors and analogs was tested at 50 µM for inhibition of nsp14 activity under conditions described for Z′-factor determination. Compounds with higher than 75% inhibition ( ) were selected for follow-up experiments. Values in a, b, and c are presented as the mean ± standard deviation of three independent experiments (n = 3). Values in d and e are from single screening.
) were selected for follow-up experiments. Values in a, b, and c are presented as the mean ± standard deviation of three independent experiments (n = 3). Values in d and e are from single screening.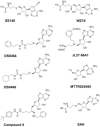
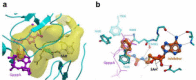
Update of
-
Probing the SAM Binding Site of SARS-CoV-2 nsp14 in vitro Using SAM Competitive Inhibitors Guides Developing Selective bi-substrate Inhibitors.bioRxiv [Preprint]. 2021 Feb 19:2021.02.19.424337. doi: 10.1101/2021.02.19.424337. bioRxiv. 2021. Update in: SLAS Discov. 2021 Oct;26(9):1200-1211. doi: 10.1177/24725552211026261. PMID: 33619486 Free PMC article. Updated. Preprint.
References
-
- Zaki A.M., van Boheemen S., Bestebroer T.M., et al. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

