An epidemic Zika virus isolate suppresses antiviral immunity by disrupting antigen presentation pathways
- PMID: 34193875
- PMCID: PMC8245533
- DOI: 10.1038/s41467-021-24340-0
An epidemic Zika virus isolate suppresses antiviral immunity by disrupting antigen presentation pathways
Abstract
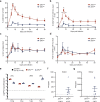
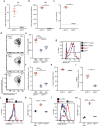
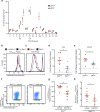
Zika virus (ZIKV) has emerged as an important global health threat, with the recently acquired capacity to cause severe neurological symptoms and to persist within host tissues. We previously demonstrated that an early Asian lineage ZIKV isolate induces a highly activated CD8 T cell response specific for an immunodominant epitope in the ZIKV envelope protein in wild-type mice. Here we show that a contemporary ZIKV isolate from the Brazilian outbreak severely limits CD8 T cell immunity in mice and blocks generation of the immunodominant CD8 T cell response. This is associated with a more sustained infection that is cleared between 7- and 14-days post-infection. Mechanistically, we demonstrate that infection with the Brazilian ZIKV isolate reduces the cross-presentation capacity of dendritic cells and fails to fully activate the immunoproteasome. Thus, our study provides an isolate-specific mechanism of host immune evasion by one Brazilian ZIKV isolate, which differs from the early Asian lineage isolate and provides potential insight into viral persistence associated with recent ZIKV outbreaks.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Japanese Encephalitis Virus Vaccination Elicits Cross-Reactive HLA-Class I-Restricted CD8 T Cell Response Against Zika Virus Infection.Front Immunol. 2020 Sep 25;11:577546. doi: 10.3389/fimmu.2020.577546. eCollection 2020. Front Immunol. 2020. PMID: 33101303 Free PMC article.
-
Analysis of the T Cell Response to Zika Virus and Identification of a Novel CD8+ T Cell Epitope in Immunocompetent Mice.PLoS Pathog. 2017 Feb 23;13(2):e1006184. doi: 10.1371/journal.ppat.1006184. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28231312 Free PMC article.
-
CD8+ T Cell Immune Response in Immunocompetent Mice during Zika Virus Infection.J Virol. 2017 Oct 27;91(22):e00900-17. doi: 10.1128/JVI.00900-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28835502 Free PMC article.
-
Protective to a T: The Role of T Cells during Zika Virus Infection.Cells. 2019 Aug 3;8(8):820. doi: 10.3390/cells8080820. Cells. 2019. PMID: 31382545 Free PMC article. Review.
-
Taking the defensive: Immune control of Zika virus infection.Virus Res. 2018 Aug 2;254:21-26. doi: 10.1016/j.virusres.2017.08.018. Epub 2017 Sep 1. Virus Res. 2018. PMID: 28867493 Free PMC article. Review.
Cited by
-
An Epidemic Zika Virus Isolate Drives Enhanced T Follicular Helper Cell and B Cell-Mediated Immunity.J Immunol. 2022 Apr 1;208(7):1719-1728. doi: 10.4049/jimmunol.2100049. Epub 2022 Mar 28. J Immunol. 2022. PMID: 35346966 Free PMC article.
-
Zika but not Dengue virus infection limits NF-κB activity in human monocyte-derived dendritic cells and suppresses their ability to activate T cells.Nat Commun. 2025 Mar 25;16(1):2695. doi: 10.1038/s41467-025-57977-2. Nat Commun. 2025. PMID: 40133263 Free PMC article.
-
Epidemic Zika virus strains from the Asian lineage induce an attenuated fetal brain pathogenicity.Nat Commun. 2024 Dec 30;15(1):10870. doi: 10.1038/s41467-024-55155-4. Nat Commun. 2024. PMID: 39738084 Free PMC article.
-
Current Advances in Zika Vaccine Development.Vaccines (Basel). 2022 Oct 28;10(11):1816. doi: 10.3390/vaccines10111816. Vaccines (Basel). 2022. PMID: 36366325 Free PMC article. Review.
-
Accurate enumeration of pathogen-specific and virtual memory CD8 T cells after infection.J Immunol. 2025 May 1;214(5):995-1007. doi: 10.1093/jimmun/vkaf007. J Immunol. 2025. PMID: 40167212
References
-
- Mallet, H. P., Vial, A. L. & Musso, D. Bilan de l’épidémie à virus Zika survenue en Polynésie française entre octobre 2013 et mars 2014. De la description de l’épidémie aux connaissances acquises après l’évènement. Bulletin épidémiologique hebdomadaire 367–373 (2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

