The plant NADPH oxidase RBOHD is required for microbiota homeostasis in leaves
- PMID: 34194036
- PMCID: PMC7612668
- DOI: 10.1038/s41564-021-00929-5
The plant NADPH oxidase RBOHD is required for microbiota homeostasis in leaves
Abstract
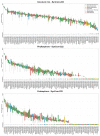
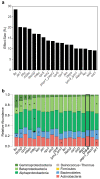
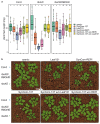
The plant microbiota consists of a multitude of microorganisms that can affect plant health and fitness. However, it is currently unclear how the plant shapes its leaf microbiota and what role the plant immune system plays in this process. Here, we evaluated Arabidopsis thaliana mutants with defects in different parts of the immune system for an altered bacterial community assembly using a gnotobiotic system. While higher-order mutants in receptors that recognize microbial features and in defence hormone signalling showed substantial microbial community alterations, the absence of the plant NADPH oxidase RBOHD caused the most pronounced change in the composition of the leaf microbiota. The rbohD knockout resulted in an enrichment of specific bacteria. Among these, we identified Xanthomonas strains as opportunistic pathogens that colonized wild-type plants asymptomatically but caused disease in rbohD knockout plants. Strain dropout experiments revealed that the lack of RBOHD unlocks the pathogenicity of individual microbiota members driving dysbiosis in rbohD knockout plants. For full protection, healthy plants require both a functional immune system and a microbial community. Our results show that the NADPH oxidase RBOHD is essential for microbiota homeostasis and emphasizes the importance of the plant immune system in controlling the leaf microbiota.
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Beattie GA, Lindow SE. Bacterial colonization of leaves: a spectrum of strategies. Phytopathol. 1999;89:353–359. - PubMed
-
- Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. - PubMed
-
- Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. - PubMed
-
- Müller DB, Vogel C, Bai Y, Vorholt JA. The plant microbiota: systems-level insights and perspectives. Annu Rev Genet. 2016;50:211–234. - PubMed
-
- Lugtenberg B, Kamilova F. Plant-Growth-Promoting Rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

