A Combined RNA Signature Predicts Recurrence Risk of Stage I-IIIA Lung Squamous Cell Carcinoma
- PMID: 34194476
- PMCID: PMC8236863
- DOI: 10.3389/fgene.2021.676464
A Combined RNA Signature Predicts Recurrence Risk of Stage I-IIIA Lung Squamous Cell Carcinoma
Abstract
Objective: Recurrence remains the main cause of the poor prognosis in stage I-IIIA lung squamous cell carcinoma (LUSC) after surgical resection. In the present study, we aimed to identify the long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and messenger RNAs (mRNAs) related to the recurrence of stage I-IIIA LUSC. Moreover, we constructed a risk assessment model to predict the recurrence of LUSC patients.
Methods: RNA sequencing data (including miRNAs, lncRNAs, and mRNAs) and relevant clinical information were obtained from The Cancer Genome Atlas (TCGA) database. The differentially expressed lncRNAs, miRNAs, and mRNAs were identified using the "DESeq2" package of the R language. Univariate Cox proportional hazards regression analysis and Kaplan-Meier curve were used to identify recurrence-related genes. Stepwise multivariate Cox regression analysis was carried out to establish a risk model for predicting recurrence in the training cohort. Moreover, Kaplan-Meier curves and receiver operating characteristic (ROC) curves were adopted to examine the predictive performance of the signature in the training cohort, validation cohort, and entire cohort.
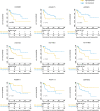
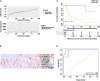
Results: Based on the TCGA database, we analyzed the differentially expressed genes (DEGs) among 27 patients with recurrent stage I-IIIA LUSC and 134 patients with non-recurrent stage I-IIIA LUSC, and identified 431 lncRNAs, 36 miRNAs, and 746 mRNAs with different expression levels. Out of these DEGs, the optimal combination of DEGs was finally determined, and a nine-joint RNA molecular signature was constructed for clinical prediction of recurrence, including LINC02683, AC244517.5, LINC02418, LINC01322, AC011468.3, hsa-mir-6825, AC020637.1, AC027117.2, and SERPINB12. The ROC curve proved that the model had good predictive performance in predicting recurrence. The area under the curve (AUC) of the prognostic model for recurrence-free survival (RFS) was 0.989 at 3 years and 0.958 at 5 years (in the training set). The combined RNA signature also revealed good predictive performance in predicting the recurrence in the validation cohort and entire cohort.
Conclusions: In the present study, we constructed a nine-joint RNA molecular signature for recurrence prediction of stage I-IIIA LUSC. Collectively, our findings provided new and valuable clinical evidence for predicting the recurrence and targeted treatment of stage I-IIIA LUSC.
Keywords: RNA signature; TCGA; biomarker; lung squamous cell carcinoma; recurrence.
Copyright © 2021 Sun, Li, Li, Yang, Zhang, Wang, Wang, Xu, Jiang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

