Human papilloma virus circulating tumor DNA assay predicts treatment response in recurrent/metastatic head and neck squamous cell carcinoma
- PMID: 34194620
- PMCID: PMC8238244
- DOI: 10.18632/oncotarget.27992
Human papilloma virus circulating tumor DNA assay predicts treatment response in recurrent/metastatic head and neck squamous cell carcinoma
Abstract
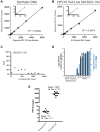
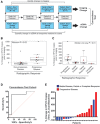
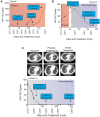
Despite the rising incidence of human papillomavirus related (HPV+) oropharyngeal squamous cell carcinoma (OPSCC), treatment of metastatic disease remains palliative. Even with new treatments such as immunotherapy, response rates are low and can be delayed, while even mild tumor progression in the face of an ineffective therapy can lead to rapid death. Real-time biomarkers of response to therapy could improve outcomes by guiding early change of therapy in the metastatic setting. Herein, we developed and analytically validated a new droplet digital PCR (ddPCR)-based assay for HPV16 circulating tumor DNA (ctDNA) and evaluated plasma HPV16 ctDNA for predicting treatment response in metastatic HPV+ OPSCC. We found that longitudinal changes HPV16 ctDNA correlate with treatment response and that ctDNA responses are observed earlier than conventional imaging (average 70 days, range: 35-166). With additional validation in multi-site studies, this assay may enable early identification of treatment failure, allowing patients to be directed promptly toward clinical trials or alternative therapies.
Keywords: HPV; circulating tumor DNA; ctDNA; head and neck cancer; oropharyngeal cancer.
Copyright: © 2021 Haring et al.
Conflict of interest statement
CONFLICTS OF INTEREST The study team has filed an invention disclosure on the HPV16 ctDNA assay and the University of Michigan intends to file a patent on the assay technology.
Figures


References
-
- Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, Peters G, Lee DJ, Leaf A, et al.. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003; 349:2091–98. 10.1056/NEJMoa031317. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

