A minimal hybridization chain reaction (HCR) system using peptide nucleic acids
- PMID: 34194712
- PMCID: PMC8208298
- DOI: 10.1039/d1sc01269j
A minimal hybridization chain reaction (HCR) system using peptide nucleic acids
Abstract
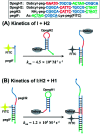
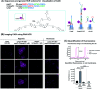
The HCR represents a powerful tool for amplification in DNA-based circuitry and sensing applications, yet requires the use of long DNA sequences to grant hairpin metastability. Here we describe a minimal HCR system based on peptide nucleic acids (PNAs). A system comprising a 5-mer stem and 5-mer loop/toehold hairpins was found to be suitable to achieve rapid amplification. These hairpins were shown to yield >10-fold amplification in 2 h and be suitable for the detection of a cancer biomarker on live cells. The use of γ-peg-modified PNA was found to be beneficial.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
A new photoelectrochemical biosensor for ultrasensitive determination of nucleic acids based on a three-stage cascade signal amplification strategy.Analyst. 2018 Jun 11;143(12):2799-2806. doi: 10.1039/c8an00609a. Analyst. 2018. PMID: 29862398
-
Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine.Chem Soc Rev. 2017 Jul 17;46(14):4281-4298. doi: 10.1039/c7cs00055c. Chem Soc Rev. 2017. PMID: 28573275 Review.
-
Novel and sensitive electrochemical/fluorescent dual-mode biosensing platform based on the cascaded cyclic amplification of enzyme-free DDSA and functional nucleic acids.Biosens Bioelectron. 2022 Dec 15;218:114762. doi: 10.1016/j.bios.2022.114762. Epub 2022 Sep 29. Biosens Bioelectron. 2022. PMID: 36195033
-
Three-dimensional DNA nanostructures to improve the hyperbranched hybridization chain reaction.Chem Sci. 2019 Aug 29;10(42):9758-9767. doi: 10.1039/c9sc02281c. eCollection 2019 Nov 14. Chem Sci. 2019. PMID: 32055345 Free PMC article.
-
Nonlinear hybridization chain reaction-based functional DNA nanostructure assembly for biosensing, bioimaging applications.Biosens Bioelectron. 2021 Feb 1;173:112814. doi: 10.1016/j.bios.2020.112814. Epub 2020 Nov 10. Biosens Bioelectron. 2021. PMID: 33197767 Review.
Cited by
-
Perspectives on conformationally constrained peptide nucleic acid (PNA): insights into the structural design, properties and applications.RSC Chem Biol. 2022 Mar 18;3(6):648-697. doi: 10.1039/d2cb00017b. eCollection 2022 Jun 8. RSC Chem Biol. 2022. PMID: 35755191 Free PMC article. Review.
-
Development of a Novel Tissue Blot Hybridization Chain Reaction for the Identification of Plant Viruses.Plants (Basel). 2022 Sep 5;11(17):2325. doi: 10.3390/plants11172325. Plants (Basel). 2022. PMID: 36079706 Free PMC article.
-
The Amplified DNA Logic Gates Based on Aptamer-Receptor Recognition for Cell Detection and Bioimaging.Biosensors (Basel). 2023 Nov 6;13(11):968. doi: 10.3390/bios13110968. Biosensors (Basel). 2023. PMID: 37998143 Free PMC article.
-
Assembly of adaptive engineering aptamers for Escherichia coli and their application in all-in-one rapid detection.Mikrochim Acta. 2025 Apr 1;192(4):272. doi: 10.1007/s00604-025-07138-5. Mikrochim Acta. 2025. PMID: 40167799
-
Impact of charges on the hybridization kinetics and thermal stability of PNA duplexes.Org Biomol Chem. 2024 Jul 17;22(28):5759-5767. doi: 10.1039/d4ob00887a. Org Biomol Chem. 2024. PMID: 38920402 Free PMC article.
References
-
- Watson E. E. Angerani S. Sabale P. M. Winssinger N. J. Am. Chem. Soc. 2021;143:4467–4482. - PubMed
-
- Yin P. Choi H. M. T. Calvert C. R. Pierce N. A. Nature. 2008;451:318–322. - PubMed
-
- Zhang D. Y. Seelig G. Nat. Chem. 2011;3:103–113. - PubMed
-
- Gorska K. Winssinger N. Angew. Chem., Int. Ed. 2013;52:6820–6843. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

