Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota
- PMID: 34194833
- PMCID: PMC8240117
- DOI: 10.1016/j.jare.2020.12.008
Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota
Abstract
Introduction: The seed-associated microbiome has a strong influence on plant ecology, fitness, and productivity. Plant microbiota could be exploited for a more responsible crop management in sustainable agriculture. However, the relationships between seed microbiota and hosts related to the changes from ancestor species to breeded crops still remain poor understood.
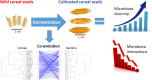
Objectives: Our aims were i) to understand the effect of cereal domestication on seed endophytes in terms of diversity, structure and co-occurrence, by comparing four cereal crops and the respective ancestor species; ii) to test the phylogenetic coherence between cereals and their seed microbiota (clue of co-evolution).
Methods: We investigated the seed microbiota of four cereal crops (Triticum aestivum, Triticum monococcum, Triticum durum, and Hordeum vulgare), along with their respective ancestors (Aegilops tauschii, Triticum baeoticum, Triticum dicoccoides, and Hordeum spontaneum, respectively) using 16S rRNA gene metabarcoding, Randomly Amplified Polymorphic DNA (RAPD) profiling of host plants and co-evolution analysis.
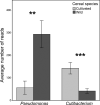
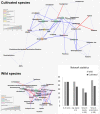
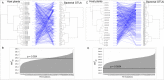
Results: The diversity of seed microbiota was generally higher in cultivated cereals than in wild ancestors, suggesting that domestication lead to a bacterial diversification. On the other hand, more microbe-microbe interactions were detected in wild species, indicating a better-structured, mature community. Typical human-associated taxa, such as Cutibacterium, dominated in cultivated cereals, suggesting an interkingdom transfers of microbes from human to plants during domestication. Co-evolution analysis revealed a significant phylogenetic congruence between seed endophytes and host plants, indicating clues of co-evolution between hosts and seed-associated microbes during domestication.
Conclusion: This study demonstrates a diversification of the seed microbiome as a consequence of domestication, and provides clues of co-evolution between cereals and their seed microbiota. This knowledge is useful to develop effective strategies of microbiome exploitation for sustainable agriculture.
Keywords: 16S metabarcoding; Cereals; Co-evolution; Domestication; Random Amplified Polymorphic DNA - RAPD; Seed microbiome.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Hardoim P.R., van Overbeek L.S., van Elsas J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. - PubMed
-
- Díaz Herrera S., Grossi C., Zawoznik M., Groppa M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol Res. 2016;186–187:37–48. - PubMed
-
- Rahman M.M., Flory E., Koyro H.W., Abideen Z., Schikora A., Suarez C. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.) Syst Appl Microbiol. 2018;41:386–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

