Determining the 3D genome structure of a single mammalian cell with Dip-C
- PMID: 34195675
- PMCID: PMC8225968
- DOI: 10.1016/j.xpro.2021.100622
Determining the 3D genome structure of a single mammalian cell with Dip-C
Abstract
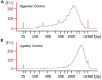
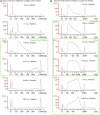
3D genome structure is highly heterogeneous among single cells and contributes to cellular functions. Our single-cell chromatin conformation capture (3C/Hi-C) technique, Dip-C, enables high-resolution (20 kb or ∼100 nm) 3D genome structure determination from single human and mouse cells. Dip-C is robust, fast, cheap, and does not require specialized equipment. This protocol describes using human and mouse brain samples to perform Dip-C, which has also been applied to other tissue types including the human blood and mouse eye, nose, and embryo. For complete details on the use and execution of this protocol, please refer to Tan et al. (2021).
Keywords: Bioinformatics; Cell isolation; Flow Cytometry/Mass Cytometry; Genetics; Genomics; Molecular Biology; Neuroscience; Sequence analysis; Sequencing; Single Cell.
© 2021 The Author(s).
Conflict of interest statement
L.T. is an inventor on a patent application (US16/615,872) filed by Harvard that covers Dip-C.
Figures





References
-
- Huang L., Ma F., Chapman A., Lu S., Xie X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu. Rev. Genomics Hum. Genet. 2015;16:79–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

