Continental-scale genomic analysis suggests shared post-admixture adaptation in the Americas
- PMID: 34196708
- PMCID: PMC8561420
- DOI: 10.1093/hmg/ddab177
Continental-scale genomic analysis suggests shared post-admixture adaptation in the Americas
Abstract
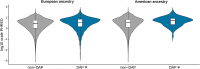
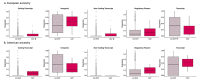
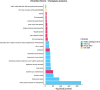
American populations are one of the most interesting examples of recently admixed groups, where ancestral components from three major continental human groups (Africans, Eurasians and Native Americans) have admixed within the last 15 generations. Recently, several genetic surveys focusing on thousands of individuals shed light on the geography, chronology and relevance of these events. However, even though gene flow could drive adaptive evolution, it is unclear whether and how natural selection acted on the resulting genetic variation in the Americas. In this study, we analysed the patterns of local ancestry of genomic fragments in genome-wide data for ~ 6000 admixed individuals from 10 American countries. In doing so, we identified regions characterized by a divergent ancestry profile (DAP), in which a significant over or under ancestral representation is evident. Our results highlighted a series of genomic regions with DAPs associated with immune system response and relevant medical traits, with the longest DAP region encompassing the human leukocyte antigen locus. Furthermore, we found that DAP regions are enriched in genes linked to cancer-related traits and autoimmune diseases. Then, analysing the biological impact of these regions, we showed that natural selection could have acted preferentially towards variants located in coding and non-coding transcripts and characterized by a high deleteriousness score. Taken together, our analyses suggest that shared patterns of post admixture adaptation occurred at a continental scale in the Americas, affecting more often functional and impactful genomic variants.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Ongaro, L., Scliar, M.O., Flores, R., Raveane, A., Marnetto, D., Sarno, S., Gnecchi-Ruscone, G.A., Alarcón-Riquelme, M.E., Patin, E., Wangkumhang, P. et al. (2019) The genomic impact of European colonization of the Americas. Curr. Biol., 29, 3974–3986.e4. - PubMed
-
- Chacón-Duque, J.-C., Adhikari, K., Fuentes-Guajardo, M., Mendoza-Revilla, J., Acuña-Alonzo, V., Barquera, R., Quinto-Sánchez, M., Gómez-Valdés, J., Everardo Martínez, P., Villamil-Ramírez, H. et al. (2018) Latin Americans show wide-spread Converso ancestry and imprint of local native ancestry on physical appearance. Nat. Commun., 9, 5388. - PMC - PubMed

