Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis
- PMID: 34197182
- PMCID: PMC8331063
- DOI: 10.1200/JCO.20.03238
Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis
Abstract
Purpose: To analyze the prevalence of homologous recombination deficiency (HRD) in patients with pancreatic ductal adenocarcinoma (PDAC).
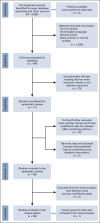
Materials and methods: We conducted a systematic review and meta-analysis of the prevalence of HRD in PDAC from PubMed, Scopus, and Cochrane Library databases, and online cancer genomic data sets. The main outcome was pooled prevalence of somatic and germline mutations in the better characterized HRD genes (BRCA1, BRCA2, PALB2, ATM, ATR, CHEK2, RAD51, and the FANC genes). The secondary outcomes were prevalence of germline mutations overall, and in sporadic and familial cases; prevalence of germline BRCA1/2 mutations in Ashkenazi Jewish (AJ); and prevalence of HRD based on other definitions (ie, alterations in other genes, genomic scars, and mutational signatures). Random-effects modeling with the Freeman-Tukey transformation was used for the analyses. PROSPERO registration number: (CRD42020190813).
Results: Sixty studies with 21,842 participants were included in the systematic review and 57 in the meta-analysis. Prevalence of germline and somatic mutations was BRCA1: 0.9%, BRCA2: 3.5%, PALB2: 0.2%, ATM: 2.2%, CHEK2: 0.3%, FANC: 0.5%, RAD51: 0.0%, and ATR: 0.1%. Prevalence of germline mutations was BRCA1: 0.9% (2.4% in AJ), BRCA2: 3.8% (8.2% in AJ), PALB2: 0.2%, ATM: 2%, CHEK2: 0.3%, and FANC: 0.4%. No significant differences between sporadic and familial cases were identified. HRD prevalence ranged between 14.5%-16.5% through targeted next-generation sequencing and 24%-44% through whole-genome or whole-exome sequencing allowing complementary genomic analysis, including genomic scars and other signatures (surrogate markers of HRD).
Conclusion: Surrogate readouts of HRD identify a greater proportion of patients with HRD than analyses limited to gene-level approaches. There is a clear need to harmonize HRD definitions and to validate the optimal biomarker for treatment selection. Universal HRD screening including integrated somatic and germline analysis should be offered to all patients with PDAC.
Conflict of interest statement
Figures

References
-
- Kamisawa T Wood LD Itoi T, et al. : Pancreatic cancer. Lancet 388:73-85, 2016 - PubMed
-
- Bray F Ferlay J Soerjomataram I, et al. : Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394-424, 2018 - PubMed
-
- Carioli G Malvezzi M Bertuccio P, et al. : European cancer mortality predictions for the year 2021 with focus on pancreatic and female lung cancer. Ann Oncol 32:478-487, 2021 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

