Dynamics of the SARS-CoV-2 nucleoprotein N-terminal domain triggers RNA duplex destabilization
- PMID: 34197802
- PMCID: PMC8239202
- DOI: 10.1016/j.bpj.2021.06.003
Dynamics of the SARS-CoV-2 nucleoprotein N-terminal domain triggers RNA duplex destabilization
Abstract
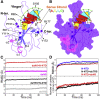
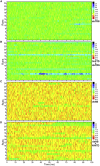
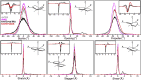
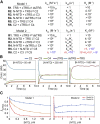
The nucleocapsid (N) protein of betacoronaviruses is responsible for nucleocapsid assembly and other essential regulatory functions. The N protein N-terminal domain (N-NTD) interacts and melts the double-stranded transcriptional regulatory sequences (dsTRSs), regulating the discontinuous subgenome transcription process. Here, we used molecular dynamics (MD) simulations to study the binding of the severe acute respiratory syndrome coronavirus 2 N-NTD to nonspecific (NS) and TRS dsRNAs. We probed dsRNAs' Watson-Crick basepairing over 25 replicas of 100 ns MD simulations, showing that only one N-NTD of dimeric N is enough to destabilize dsRNAs, triggering melting initiation. dsRNA destabilization driven by N-NTD was more efficient for dsTRSs than dsNS. N-NTD dynamics, especially a tweezer-like motion of β2-β3 and Δ2-β5 loops, seems to play a key role in Watson-Crick basepairing destabilization. Based on experimental information available in the literature, we constructed kinetics models for N-NTD-mediated dsRNA melting. Our results support a 1:1 stoichiometry (N-NTD/dsRNA), matching MD simulations and raising different possibilities for N-NTD action: 1) two N-NTD arms of dimeric N would bind to two different RNA sites, either closely or spatially spaced in the viral genome, in a cooperative manner; and 2) monomeric N-NTD would be active, opening up the possibility of a regulatory dissociation event.
Copyright © 2021. Published by Elsevier Inc.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

