Epigenetic Deregulation of Apoptosis in Cancers
- PMID: 34199020
- PMCID: PMC8267644
- DOI: 10.3390/cancers13133210
Epigenetic Deregulation of Apoptosis in Cancers
Abstract
Cancer cells possess the ability to evade apoptosis. Genetic alterations through mutations in key genes of the apoptotic signaling pathway represent a major adaptive mechanism of apoptosis evasion. In parallel, epigenetic changes via aberrant modifications of DNA and histones to regulate the expression of pro- and antiapoptotic signal mediators represent a major complementary mechanism in apoptosis regulation and therapy response. Most epigenetic changes are governed by the activity of chromatin modifying enzymes that add, remove, or recognize different marks on histones and DNA. Here, we discuss how apoptosis signaling components are deregulated at epigenetic levels, particularly focusing on the roles of chromatin-modifying enzymes in this process. We also review the advances in cancer therapies with epigenetic drugs such as DNMT, HMT, HDAC, and BET inhibitors, as well as their effects on apoptosis modulation in cancer cells. Rewiring the epigenome by drug interventions can provide therapeutic advantage for various cancers by reverting therapy resistance and leading cancer cells to undergo apoptotic cell death.
Keywords: apoptosis; cancer; chromatin modifying enzymes; epigenetic drugs; evasion; therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

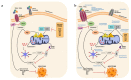
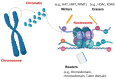


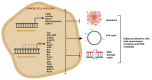

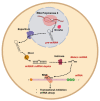
References
-
- Green D.R. Means to an End: Apoptosis and Other Cell Death Mechanisms. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2010.
Publication types
LinkOut - more resources
Full Text Sources

