Post Zygotic, Somatic, Deletion in KERATIN 1 V1 Domain Generates Structural Alteration of the K1/K10 Dimer, Producing a Monolateral Palmar Epidermolytic Nevus
- PMID: 34199056
- PMCID: PMC8269197
- DOI: 10.3390/ijms22136901
Post Zygotic, Somatic, Deletion in KERATIN 1 V1 Domain Generates Structural Alteration of the K1/K10 Dimer, Producing a Monolateral Palmar Epidermolytic Nevus
Abstract
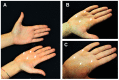
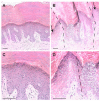
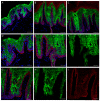
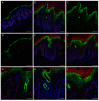
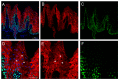
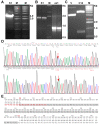
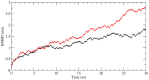
Palmoplantar keratodermas (PPKs) are characterized by thickness of stratum corneum and epidermal hyperkeratosis localized in palms and soles. PPKs can be epidermolytic (EPPK) or non epidermolytic (NEPPK). Specific mutations of keratin 16 (K16) and keratin 1 (K1) have been associated to EPPK, and NEPPK. Cases of mosaicism in PPKs due to somatic keratin mutations have also been described in scientific literature. We evaluated a patient presenting hyperkeratosis localized monolaterally in the right palmar area, characterized by linear yellowish hyperkeratotic lesions following the Blaschko lines. No other relatives of the patient showed any dermatological disease. Light and confocal histological analysis confirmed the presence of epidermolityic hyperkeratosis. Genetic analysis performed demonstrates the heterozygous deletion NM_006121.4:r.274_472del for a total of 198 nucleotides, in KRT1 cDNA obtained by a palmar lesional skin biopsy, corresponding to the protein mutation NP_006112.3:p.Gly71_Gly137del. DNA extracted from peripheral blood lymphocytes did not display the presence of the mutation. These results suggest a somatic mutation causing an alteration in K1 N-terminal variable domain (V1). The deleted sequence involves the ISIS subdomain, containing a lysine residue already described as fundamental for epidermal transglutaminases in the crosslinking of IF cytoskeleton. Moreover, a computational analysis of the wild-type and V1-mutated K1/K10 keratin dimers, suggests an unusual interaction between these keratin filaments. The mutation taster in silico analysis also returned a high probability for a deleterious mutation. These data demonstrate once again the importance of the head domain (V1) of K1 in the formation of a functional keratinocyte cytoskeleton. Moreover, this is a further demonstration of the presence of somatic mutations arising in later stages of the embryogenesis, generating a mosaic phenotype.
Keywords: EPPK; KIF; NEPPK; genodermatosis; keratin structure; keratins; mosaic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
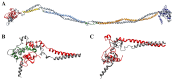
Similar articles
-
Keratin 1 gene mutation detected in epidermal nevus with epidermolytic hyperkeratosis.J Invest Dermatol. 2007 Jun;127(6):1371-4. doi: 10.1038/sj.jid.5700712. Epub 2007 Feb 1. J Invest Dermatol. 2007. PMID: 17255957
-
Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.EMBO J. 2019 Jun 3;38(11):e100741. doi: 10.15252/embj.2018100741. Epub 2019 Apr 29. EMBO J. 2019. PMID: 31036554 Free PMC article.
-
Atypical epidermolytic palmoplantar keratoderma presentation associated with a mutation in the keratin 1 gene.Br J Dermatol. 2004 Jun;150(6):1096-103. doi: 10.1111/j.1365-2133.2004.05967.x. Br J Dermatol. 2004. PMID: 15214894
-
Human keratin diseases: hereditary fragility of specific epithelial tissues.Exp Dermatol. 1996 Dec;5(6):297-307. doi: 10.1111/j.1600-0625.1996.tb00133.x. Exp Dermatol. 1996. PMID: 9028791 Review.
-
Keratin 9 L164P mutation in a Chinese pedigree with epidermolytic palmoplantar keratoderma, cytokeratin analysis, and literature review.Mol Genet Genomic Med. 2019 Nov;7(11):e977. doi: 10.1002/mgg3.977. Epub 2019 Sep 16. Mol Genet Genomic Med. 2019. PMID: 31525823 Free PMC article. Review.
Cited by
-
Anlotinib plus icotinib as a potential treatment option for EGFR-mutated advanced non-squamous non-small cell lung cancer with concurrent mutations: final analysis of the prospective phase 2, multicenter ALTER-L004 study.Mol Cancer. 2023 Aug 5;22(1):124. doi: 10.1186/s12943-023-01823-w. Mol Cancer. 2023. PMID: 37543587 Free PMC article. Clinical Trial.
-
Unveiling the impact of hypodermis on gene expression for advancing bioprinted full-thickness 3D skin models.Commun Biol. 2024 Nov 11;7(1):1437. doi: 10.1038/s42003-024-07106-4. Commun Biol. 2024. PMID: 39528562 Free PMC article.
References
-
- Oji V., Tadini G., Akiyama M., Bardon C.B., Bodemer C., Bourrat E., Coudiere P., DiGiovanna J.J., Elias P., Fischer J., et al. Revised nomenclature and classification of inherited ichthyoses: Results of the First Ichthyosis Consensus Conference in Sorèze 2009. J. Am. Acad. Dermatol. 2010;63:607–641. doi: 10.1016/j.jaad.2009.11.020. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

