Functions of Coronavirus Accessory Proteins: Overview of the State of the Art
- PMID: 34199223
- PMCID: PMC8231932
- DOI: 10.3390/v13061139
Functions of Coronavirus Accessory Proteins: Overview of the State of the Art
Abstract
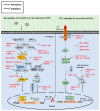
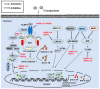
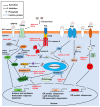
Coronavirus accessory proteins are a unique set of proteins whose genes are interspersed among or within the genes encoding structural proteins. Different coronavirus genera, or even different species within the same coronavirus genus, encode varying amounts of accessory proteins, leading to genus- or species-specificity. Though accessory proteins are dispensable for the replication of coronavirus in vitro, they play important roles in regulating innate immunity, viral proliferation, and pathogenicity. The function of accessory proteins on virus infection and pathogenesis is an area of particular interest. In this review, we summarize the current knowledge on accessory proteins of several representative coronaviruses that infect humans or animals, including the emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with an emphasis on their roles in interaction between virus and host, mainly involving stress response, innate immunity, autophagy, and apoptosis. The cross-talking among these pathways is also discussed.
Keywords: accessory protein; coronavirus; infection; innate immunity; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection.mBio. 2019 Mar 26;10(2):e00319-19. doi: 10.1128/mBio.00319-19. mBio. 2019. PMID: 30914508 Free PMC article.
-
Involvement of SARS-CoV-2 accessory proteins in immunopathogenesis.Microbiol Immunol. 2024 Jul;68(7):237-247. doi: 10.1111/1348-0421.13157. Epub 2024 Jun 4. Microbiol Immunol. 2024. PMID: 38837257
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
-
SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns.Front Immunol. 2021 Jul 7;12:708264. doi: 10.3389/fimmu.2021.708264. eCollection 2021. Front Immunol. 2021. PMID: 34305949 Free PMC article. Review.
-
The N-Terminal Region of Middle East Respiratory Syndrome Coronavirus Accessory Protein 8b Is Essential for Enhanced Virulence of an Attenuated Murine Coronavirus.J Virol. 2022 Feb 9;96(3):e0184221. doi: 10.1128/JVI.01842-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34817197 Free PMC article.
Cited by
-
A peptide array pipeline for the development of Spike-ACE2 interaction inhibitors.Peptides. 2022 Dec;158:170898. doi: 10.1016/j.peptides.2022.170898. Epub 2022 Oct 21. Peptides. 2022. PMID: 36279985 Free PMC article.
-
The Regulatory Network of Cyclic GMP-AMP Synthase-Stimulator of Interferon Genes Pathway in Viral Evasion.Front Microbiol. 2021 Dec 13;12:790714. doi: 10.3389/fmicb.2021.790714. eCollection 2021. Front Microbiol. 2021. PMID: 34966372 Free PMC article. Review.
-
The role of SARS-CoV-2 accessory proteins in immune evasion.Biomed Pharmacother. 2022 Dec;156:113889. doi: 10.1016/j.biopha.2022.113889. Epub 2022 Oct 17. Biomed Pharmacother. 2022. PMID: 36265309 Free PMC article. Review.
-
Infection of primary nasal epithelial cells differentiates among lethal and seasonal human coronaviruses.bioRxiv [Preprint]. 2022 Oct 18:2022.10.17.512617. doi: 10.1101/2022.10.17.512617. bioRxiv. 2022. Update in: Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2218083120. doi: 10.1073/pnas.2218083120. PMID: 36299422 Free PMC article. Updated. Preprint.
-
Severe acute respiratory syndrome coronaviruses contributing to mitochondrial dysfunction: Implications for post-COVID complications.Mitochondrion. 2023 Mar;69:43-56. doi: 10.1016/j.mito.2023.01.005. Epub 2023 Jan 20. Mitochondrion. 2023. PMID: 36690315 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

