Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression
- PMID: 34199780
- PMCID: PMC8230009
- DOI: 10.3390/brainsci11060746
Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression
Abstract
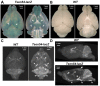
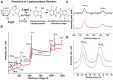
Acquisition of detailed anatomical and molecular knowledge from intact biological samples while preserving their native three-dimensional structure is still a challenging issue for imaging studies aiming to unravel a system's functions. Three-dimensional micro-CT X-ray imaging with a high spatial resolution in minimally perturbed naive non-transparent samples has recently gained increased popularity and broad application in biomedical research. Here, we describe a novel X-ray-based methodology for analysis of β-galactosidase (lacZ) reporter-driven gene expression in an intact murine brain ex vivo by micro-CT. The method relies on detection of bromine molecules in the product of the enzymatic β-galactosidase reaction. Enhancement of the X-ray signal is observed specifically in the regions of the murine brain where expression of the lacZ reporter gene is also detected histologically. We performed quantitative analysis of the expression levels of lacZ reporter activity by relative radiodensity estimation of the β-galactosidase/X-gal precipitate in situ. To demonstrate the feasibility of the method, we performed expression analysis of the Tsen54-lacZ reporter gene in the murine brain in a semi-quantitative manner. Human mutations in the Tsen54 gene cause pontocerebellar hypoplasia (PCH), a group of severe neurodegenerative disorders with both mental and motor deficits. Comparing relative levels of Tsen54 gene expression, we demonstrate that the highest Tsen54 expression is observed in anatomical brain substructures important for the normal motor and memory functions in mice.
Keywords: Tsen54; X-ray imaging; gene expression; lacZ reporter; mouse brain; mouse phenotyping; pontocerebellar hypoplesia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

