Characterization of Rheumatoid Arthritis Risk-Associated SNPs and Identification of Novel Therapeutic Sites Using an In-Silico Approach
- PMID: 34199962
- PMCID: PMC8227790
- DOI: 10.3390/biology10060501
Characterization of Rheumatoid Arthritis Risk-Associated SNPs and Identification of Novel Therapeutic Sites Using an In-Silico Approach
Abstract
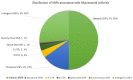
Single-nucleotide polymorphisms (SNPs) are reported to be associated with many diseases, including autoimmune diseases. In rheumatoid arthritis (RA), about 152 SNPs are reported to account for ~15% of its heritability. These SNPs may result in the alteration of gene expression and may also affect the stability of mRNA, resulting in diseased protein. Therefore, in order to predict the underlying mechanism of these SNPs and identify novel therapeutic sites for the treatment of RA, several bioinformatics tools were used. The damaging effect of 23 non-synonymous SNPs on proteins using different tools suggested four SNPs, including rs2476601 in PTPN22, rs5029941 and rs2230926 in TNFAIP3, and rs34536443 in TYK2, to be the most damaging. In total, 42 of 76 RA-associated intronic SNPs were predicted to create or abolish potential splice sites. Moreover, the analysis of 11 RA-associated UTR SNPs indicated that only one SNP, rs1128334, located in 3'UTR of ETS1, caused functional pattern changes in BRD-BOX. For the identification of novel therapeutics sites to treat RA, extensive gene-gene interaction network interactive pathways were established, with the identification of 13 potential target sites for the development of RA drugs, including three novel target genes. The anticipated effect of these findings on RA pathogenesis may be further validated in both in vivo and in vitro studies.
Keywords: SNPs; gene–gene interaction; miRNA; rheumatoid arthritis; therapeutic sites.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Aho K., Koskenvuo M., Tuominen J., Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 1986;13:899–902. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

