The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol
- PMID: 34200052
- PMCID: PMC8200238
- DOI: 10.3390/molecules26113413
The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol
Abstract
A systematic investigation of the experimental 13C-NMR spectra published in Molecules during the period of 1996 to 2015 with respect to their quality using CSEARCH-technology is described. It is shown that the systematic application of the CSEARCH-Robot-Referee during the peer-reviewing process prohibits at least the most trivial assignment errors and wrong structure proposals. In many cases, the correction of the assignments/chemical shift values is possible by manual inspection of the published tables; in certain cases, reprocessing of the original experimental data might help to clarify the situation, showing the urgent need for a public domain repository. A comparison of the significant key numbers derived for Molecules against those of other important journals in the field of natural product chemistry shows a quite similar level of quality for all publishers responsible for the six journals under investigation. From the results of this study, general rules for data handling, data storage, and manuscript preparation can be derived, helping to increase the quality of published NMR-data and making these data available as validated reference material.
Keywords: 13C-NMR; NMR; computer-assisted peer-reviewing; database; spectrum prediction; structure generation.
Conflict of interest statement
The author declares no conflict of interest.
Figures
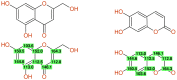


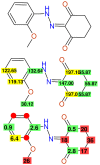
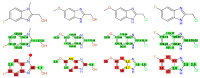
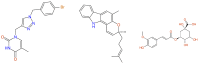





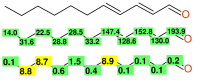

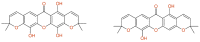
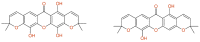


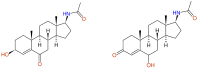



References
-
- McAlpine J.B., Chen S.N., Kutateladze A., MacMillan J.B., Appendino G., Barison A., Beniddir M.A., Biavatti M.W., Bluml S., Boufridi A., et al. The value of universally available raw NMR data for transparency, reproducibility, and integrity in natural product research. Nat. Prod. Rep. 2019;36:35–107. doi: 10.1039/C7NP00064B. - DOI - PMC - PubMed
-
- Robien W. A Critical Evaluation of the Quality of Published 13C NMR Data in Natural Product Chemistry. In: Kinghorn A.D., Falk H., Gibbons S., Kobayashi J., editors. Progress in the Chemistry of Organic Natural Products. Volume 105. Springer International Publishing; Cham, Switzerland: 2017. pp. 137–215. - DOI - PubMed
-
- Bremser W. HOSE—A novel substructure code. Anal. Chim. Acta. 1978;103:355–365. doi: 10.1016/S0003-2670(01)83100-7. - DOI
-
- Purtuc V. Master’s Thesis. University of Vienna; Vienna, Austria: 1997. Abschätzung von 13C-NMR-Spektren Mittels Neuronaler Netze.
LinkOut - more resources
Full Text Sources

