Grouper TRAF4, a Novel, CP-Interacting Protein That Promotes Red-Spotted Grouper Nervous Necrosis Virus Replication
- PMID: 34200212
- PMCID: PMC8201248
- DOI: 10.3390/ijms22116136
Grouper TRAF4, a Novel, CP-Interacting Protein That Promotes Red-Spotted Grouper Nervous Necrosis Virus Replication
Abstract
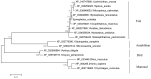
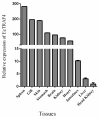
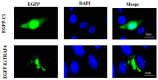
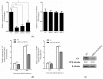
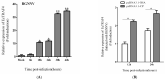
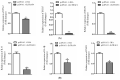
Tumor necrosis factor receptor-associated factors (TRAFs) play important roles in the biological processes of immune regulation, the inflammatory response, and apoptosis. TRAF4 belongs to the TRAF family and plays a major role in many biological processes. Compared with other TRAF proteins, the functions of TRAF4 in teleosts have been largely unknown. In the present study, the TRAF4 homologue (EcTRAF4) of the orange-spotted grouper was characterized. EcTRAF4 consisted of 1413 bp encoding a 471-amino-acid protein, and the predicted molecular mass was 54.27 kDa. EcTRAF4 shares 99.79% of its identity with TRAF4 of the giant grouper (E. lanceolatus). EcTRAF4 transcripts were ubiquitously and differentially expressed in all the examined tissues. EcTRAF4 expression in GS cells was significantly upregulated after stimulation with red-spotted grouper nervous necrosis virus (RGNNV). EcTRAF4 protein was distributed in the cytoplasm of GS cells. Overexpressed EcTRAF4 promoted RGNNV replication during viral infection in vitro. Yeast two-hybrid and coimmunoprecipitation assays showed that EcTRAF4 interacted with the coat protein (CP) of RGNNV. EcTRAF4 inhibited the activation of IFN3, IFN-stimulated response element (ISRE), and nuclear factor-κB (NF-κB). Overexpressed EcTRAF4 also reduced the expression of interferon (IFN)-related molecules and pro-inflammatory factors. Together, these results demonstrate that EcTRAF4 plays crucial roles in RGNNV infection.
Keywords: Epinephelus coioides; RGNNV; TRAF4; cellular localization; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



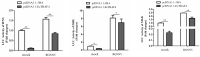
Similar articles
-
Fish RIP1 Mediates Innate Antiviral Immune Responses Induced by SGIV and RGNNV Infection.Front Immunol. 2020 Aug 4;11:1718. doi: 10.3389/fimmu.2020.01718. eCollection 2020. Front Immunol. 2020. PMID: 32849607 Free PMC article.
-
MicroRNA-146a promotes red spotted grouper nervous necrosis virus (RGNNV) replication by targeting TRAF6 in orange spotted grouper, Epinephelus coioides.Fish Shellfish Immunol. 2018 Jan;72:9-13. doi: 10.1016/j.fsi.2017.10.020. Epub 2017 Oct 24. Fish Shellfish Immunol. 2018. PMID: 29074132
-
Fish Snx27 promotes viral products by modulating the innate immune response and exosomal machinery.J Virol. 2024 Dec 17;98(12):e0097424. doi: 10.1128/jvi.00974-24. Epub 2024 Nov 4. J Virol. 2024. PMID: 39494909 Free PMC article.
-
Transducin β-like 1 X-linked receptor 1 (TBLR1) affects RGNNV infection through negative regulation of interferon immune response in orange-spotted grouper, Epinephelus coioides.Fish Shellfish Immunol. 2019 Jun;89:76-82. doi: 10.1016/j.fsi.2019.03.046. Epub 2019 Mar 24. Fish Shellfish Immunol. 2019. PMID: 30917925
-
The Research Progress in Physiological and Pathological Functions of TRAF4.Front Oncol. 2022 Feb 15;12:842072. doi: 10.3389/fonc.2022.842072. eCollection 2022. Front Oncol. 2022. PMID: 35242717 Free PMC article. Review.
Cited by
-
Transcriptome Analysis Reveals the Immunoregulatory Activity of Rice Seed-Derived Peptide PEP1 on Dendritic Cells.Molecules. 2023 Jul 5;28(13):5224. doi: 10.3390/molecules28135224. Molecules. 2023. PMID: 37446885 Free PMC article.
-
Novel Findings in Teleost TRAF4, a Protein Acts as an Enhancer in TRIF and TRAF6 Mediated Antiviral and Inflammatory Signaling.Front Immunol. 2022 Jul 11;13:944528. doi: 10.3389/fimmu.2022.944528. eCollection 2022. Front Immunol. 2022. PMID: 35898509 Free PMC article.
-
Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection.Int J Mol Sci. 2023 May 6;24(9):8363. doi: 10.3390/ijms24098363. Int J Mol Sci. 2023. PMID: 37176078 Free PMC article.
-
Dietary Administration of Novel Multistrain Probiotics from Healthy Grouper Intestines Promotes the Intestinal Immune Response against NNV Infection.Life (Basel). 2021 Oct 7;11(10):1053. doi: 10.3390/life11101053. Life (Basel). 2021. PMID: 34685424 Free PMC article.
-
Grouper TRIM23 exerts antiviral activity against iridovirus and nodavirus.Front Immunol. 2022 Sep 20;13:985291. doi: 10.3389/fimmu.2022.985291. eCollection 2022. Front Immunol. 2022. PMID: 36203610 Free PMC article.
References
-
- Zhu Z.M., Dong C.F., Weng S.P., He J.G. The high prevalence of pathogenic Vibrio harveyi with multiple antibiotic resistance in scale drop and muscle necrosis disease of the hybrid grouper, Epinephelus fuscoguttatus (female symbol) × E. lanceolatus (male symbol), in China. J. Fish Dis. 2018;41:589–601. doi: 10.1111/jfd.12758. - DOI - PubMed
-
- Hegde A., Chen C., Qin Q., Lam T., Sin Y. Characterization, pathogenicity and neutralization studies of a nervous necrosis virus isolated from grouper, Epinephelus tauvina, in Singapore. Aquaculture. 2002;213:55–72. doi: 10.1016/S0044-8486(02)00092-3. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

