Specificity of the HIV-1 Protease on Substrates Representing the Cleavage Site in the Proximal Zinc-Finger of HIV-1 Nucleocapsid Protein
- PMID: 34201134
- PMCID: PMC8227227
- DOI: 10.3390/v13061092
Specificity of the HIV-1 Protease on Substrates Representing the Cleavage Site in the Proximal Zinc-Finger of HIV-1 Nucleocapsid Protein
Abstract
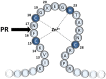
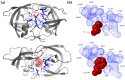
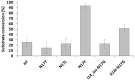
To explore the sequence context-dependent nature of the human immunodeficiency virus type 1 (HIV-1) protease's specificity and to provide a rationale for viral mutagenesis to study the potential role of the nucleocapsid (NC) processing in HIV-1 replication, synthetic oligopeptide substrates representing the wild-type and modified versions of the proximal cleavage site of HIV-1 NC were assayed as substrates of the HIV-1 protease (PR). The S1' substrate binding site of HIV-1 PR was studied by an in vitro assay using KIVKCF↓NCGK decapeptides having amino acid substitutions of N17 residue of the cleavage site of the first zinc-finger domain, and in silico calculations were also performed to investigate amino acid preferences of S1' site. Second site substitutions have also been designed to produce "revertant" substrates and convert a non-hydrolysable sequence (having glycine in place of N17) to a substrate. The specificity constants obtained for peptides containing non-charged P1' substitutions correlated well with the residue volume, while the correlation with the calculated interaction energies showed the importance of hydrophobicity: interaction energies with polar residues were related to substantially lower specificity constants. Cleavable "revertants" showed one residue shift of cleavage position due to an alternative productive binding mode, and surprisingly, a double cleavage of a substrate was also observed. The results revealed the importance of alternative binding possibilities of substrates into the HIV-1 PR. The introduction of the "revertant" mutations into infectious virus clones may provide further insights into the potential role of NC processing in the early phase of the viral life-cycle.
Keywords: HIV-1; human immunodeficiency virus; nucleocapsid protein; protease; retroviruses; specificity; substrate specificity; viral proteases; viral proteins.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

