SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2
- PMID: 34201214
- PMCID: PMC8226513
- DOI: 10.3390/cells10061434
SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2
Abstract
In late 2019, the betacoronavirus SARS-CoV-2 was identified as the viral agent responsible for the coronavirus disease 2019 (COVID-19) pandemic. Coronaviruses Spike proteins are responsible for their ability to interact with host membrane receptors and different proteins have been identified as SARS-CoV-2 interactors, among which Angiotensin-converting enzyme 2 (ACE2), and Basigin2/EMMPRIN/CD147 (CD147). CD147 plays an important role in human immunodeficiency virus type 1, hepatitis C virus, hepatitis B virus, Kaposi's sarcoma-associated herpesvirus, and severe acute respiratory syndrome coronavirus infections. In particular, SARS-CoV recognizes the CD147 receptor expressed on the surface of host cells by its nucleocapsid protein binding to cyclophilin A (CyPA), a ligand for CD147. However, the involvement of CD147 in SARS-CoV-2 infection is still debated. Interference with both the function (blocking antibody) and the expression (knock down) of CD147 showed that this receptor partakes in SARS-CoV-2 infection and provided additional clues on the underlying mechanism: CD147 binding to CyPA does not play a role; CD147 regulates ACE2 levels and both receptors are affected by virus infection. Altogether, these findings suggest that CD147 is involved in SARS-CoV-2 tropism and represents a possible therapeutic target to challenge COVID-19.
Keywords: ACE2; CD147; COVID-19; EMMPRIN; SARS-CoV-2; basigin; entry; infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
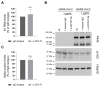
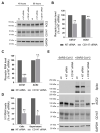
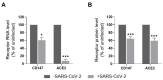
References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

