Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs
- PMID: 34201352
- PMCID: PMC8229349
- DOI: 10.3390/ijms22126200
Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs
Abstract
The VPS13 family of proteins have emerged as key players in intracellular lipid transport and human health. Humans have four different VPS13 orthologs, the dysfunction of which leads to different diseases. Yeast has a single VPS13 gene, which encodes a protein that localizes to multiple different membrane contact sites. The yeast vps13Δ mutant is pleiotropic, exhibiting defects in sporulation, protein trafficking, endoplasmic reticulum (ER)-phagy and mitochondrial function. Non-null alleles resulting from missense mutations can be useful reagents for understanding the multiple functions of a gene. The exceptionally large size of Vps13 makes the identification of key residues challenging. As a means to identify critical residues in yeast Vps13, amino acid substitution mutations from VPS13A, B, C and D, associated with human disease, were introduced at the cognate positions of yeast VPS13, some of which created separation-of-function alleles. Phenotypic analyses of these mutants have revealed that the promotion of ER-phagy is a fourth, genetically separable role of VPS13 and provide evidence that co-adaptors at the endosome mediate the activity of VPS13 in vacuolar sorting.
Keywords: ER-phagy; Vps13 adaptor; chorea-acanthocytosis; protein trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


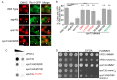
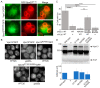

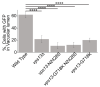

References
-
- Kolehmainen J., Black G.C., Saarinen A., Chandler K., Clayton-Smith J., Traskelin A.L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., et al. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 2003;72:1359–1369. doi: 10.1086/375454. - DOI - PMC - PubMed
-
- Lesage S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A., Cormier-Dequaire F., Hassoun S.M., Pujol C., Ciura S., et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016;98:500–513. doi: 10.1016/j.ajhg.2016.01.014. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

