Membrane Interactions Accelerate the Self-Aggregation of Huntingtin Exon 1 Fragments in a Polyglutamine Length-Dependent Manner
- PMID: 34201610
- PMCID: PMC8268948
- DOI: 10.3390/ijms22136725
Membrane Interactions Accelerate the Self-Aggregation of Huntingtin Exon 1 Fragments in a Polyglutamine Length-Dependent Manner
Abstract
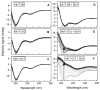
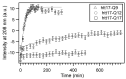
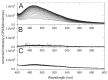
The accumulation of aggregated protein is a typical hallmark of many human neurodegenerative disorders, including polyglutamine-related diseases such as chorea Huntington. Misfolding of the amyloidogenic proteins gives rise to self-assembled complexes and fibres. The huntingtin protein is characterised by a segment of consecutive glutamines which, when exceeding ~ 37 residues, results in the occurrence of the disease. Furthermore, it has also been demonstrated that the 17-residue amino-terminal domain of the protein (htt17), located upstream of this polyglutamine tract, strongly correlates with aggregate formation and pathology. Here, we demonstrate that membrane interactions strongly accelerate the oligomerisation and β-amyloid fibril formation of htt17-polyglutamine segments. By using a combination of biophysical approaches, the kinetics of fibre formation is investigated and found to be strongly dependent on the presence of lipids, the length of the polyQ expansion, and the polypeptide-to-lipid ratio. Finally, the implications for therapeutic approaches are discussed.
Keywords: Huntington’s disease; amyloid; circular dichroism; dynamic light scattering; htt17; huntingtin; membrane-driven aggregation; peptide-lipid interactions; thioflavin T fluorescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rubinsztein D.C., Leggo J., Coles R., Almqvist E., Biancalana V., Cassiman J.J., Chotai K., Connarty M., Crauford D., Curtis A., et al. Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am. J. Hum. Genet. 1996;59:16–22. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

