Monosomy 3 Is Linked to Resistance to MEK Inhibitors in Uveal Melanoma
- PMID: 34201614
- PMCID: PMC8269285
- DOI: 10.3390/ijms22136727
Monosomy 3 Is Linked to Resistance to MEK Inhibitors in Uveal Melanoma
Abstract
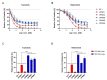
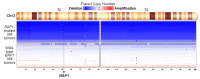
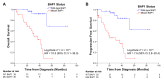
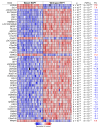
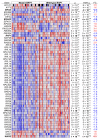
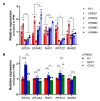
The use of MEK inhibitors in the therapy of uveal melanoma (UM) has been investigated widely but has failed to show benefits in clinical trials due to fast acquisition of resistance. In this study, we investigated a variety of therapeutic compounds in primary-derived uveal melanoma cell lines and found monosomy of chromosome 3 (M3) and mutations in BAP1 to be associated with higher resistance to MEK inhibition. However, reconstitution of BAP1 in a BAP1-deficient UM cell line was unable to restore sensitivity to MEK inhibition. We then compared UM tumors from The Cancer Genome Atlas (TCGA) with mutations in BAP1 with tumors with wild-type BAP1. Principal component analysis (PCA) clearly differentiated both groups of tumors, which displayed disparate overall and progression-free survival data. Further analysis provided insight into differential expression of genes involved in signaling pathways, suggesting that the downregulation of the eukaryotic translation initiation factor 2A (EIF2A) observed in UM tumors with BAP1 mutations and M3 UM cell lines might lead to a decrease in ribosome biogenesis while inducing an adaptive response to stress. Taken together, our study links loss of chromosome 3 with decreased sensitivity to MEK inhibition and gives insight into possible related mechanisms, whose understanding is fundamental to overcome resistance in this aggressive tumor.
Keywords: BRCA1-associated protein 1; MAPK; eIF2 signaling; eIF2alpha; monosomy 3; ocular melanoma; targeted therapy.
Conflict of interest statement
J.T.S. reports the following disclosures: Bristol-Myers Sqibb, Celgene, Roche (Research Funding); AstraZeneca, Bristol-Myers Squibb, Celgene, Immunocore, Novartis, Roche, Shire (Consulting or advisory role); AstraZeneca, Aurikamed, Baxalta, Bristol Myers Squibb, Celgene, Falk Foundation, iomedico, Immunocore, Novartis, Roche, Shire (honoraria); minor equity in FAPI Holding and Pharma15 (<3%) and member of the Board of Directors for Pharma15, outside the submitted work. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






Similar articles
-
Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma.Invest Ophthalmol Vis Sci. 2014 Jun 26;55(8):5160-7. doi: 10.1167/iovs.14-14550. Invest Ophthalmol Vis Sci. 2014. PMID: 24970262
-
Prognostic value of chromosomal imbalances, gene mutations, and BAP1 expression in uveal melanoma.Genes Chromosomes Cancer. 2018 Aug;57(8):387-400. doi: 10.1002/gcc.22541. Genes Chromosomes Cancer. 2018. PMID: 29689622
-
Comparison of Germline versus Somatic BAP1 Mutations for Risk of Metastasis in Uveal Melanoma.BMC Cancer. 2018 Nov 26;18(1):1172. doi: 10.1186/s12885-018-5079-x. BMC Cancer. 2018. PMID: 30477459 Free PMC article.
-
Establishment and characterization of two uveal melanoma cell lines derived from tumors with loss of one chromosome 3.Exp Eye Res. 2006 Oct;83(4):858-64. doi: 10.1016/j.exer.2006.04.004. Epub 2006 Jun 5. Exp Eye Res. 2006. PMID: 16750193 Review.
-
Ocular melanoma and the BAP1 hereditary cancer syndrome: implications for the dermatologist.Int J Dermatol. 2014 Jun;53(6):657-63. doi: 10.1111/ijd.12386. Epub 2014 Apr 2. Int J Dermatol. 2014. PMID: 24697775 Review.
Cited by
-
The Chick Chorioallantoic Membrane as a Xenograft Model for the Quantitative Analysis of Uveal Melanoma Metastasis in Multiple Organs.Cells. 2024 Jul 9;13(14):1169. doi: 10.3390/cells13141169. Cells. 2024. PMID: 39056751 Free PMC article.
-
Rescue of p53 functions by in vitro-transcribed mRNA impedes the growth of high-grade serous ovarian cancer.Cancer Commun (Lond). 2024 Jan;44(1):101-126. doi: 10.1002/cac2.12511. Epub 2023 Dec 22. Cancer Commun (Lond). 2024. PMID: 38140698 Free PMC article.
-
Nintedanib and Dasatinib Treatments Induce Protective Autophagy as a Potential Resistance Mechanism in MPM Cells.Front Cell Dev Biol. 2022 Mar 22;10:852812. doi: 10.3389/fcell.2022.852812. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35392170 Free PMC article.
-
Pharmacological drug screening to inhibit uveal melanoma metastatic cells either via EGF-R, MAPK, mTOR or PI3K.Int J Ophthalmol. 2022 Oct 18;15(10):1569-1576. doi: 10.18240/ijo.2022.10.02. eCollection 2022. Int J Ophthalmol. 2022. PMID: 36262851 Free PMC article.
-
Cerivastatin Synergizes with Trametinib and Enhances Its Efficacy in the Therapy of Uveal Melanoma.Cancers (Basel). 2023 Jan 31;15(3):886. doi: 10.3390/cancers15030886. Cancers (Basel). 2023. PMID: 36765842 Free PMC article.
References
-
- Bedikian A.Y., Legha S.S., Mavligit G., Carrasco C.H., Khorana S., Plager C., Papadopoulos N., Benjamin R.S. Treatment of uveal melanoma metastatic to the liver: A review of the M. D. Anderson cancer center experience and prognostic factors. Cancer. 1995;76:1665–1670. doi: 10.1002/1097-0142(19951101)76:9<1665::AID-CNCR2820760925>3.0.CO;2-J. - DOI - PubMed
-
- Diener-West M., Reynolds S.M., Agugliaro D.J., Caldwell R., Cumming K., Earle J.D., Hawkins B.S., Hayman J.A., Jaiyesimi I., Jampol L.M., et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005;123:1639–1643. doi: 10.1001/archopht.123.12.1639. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

