5' Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints
- PMID: 34202044
- PMCID: PMC8268747
- DOI: 10.3390/cancers13133171
5' Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints
Abstract
Background: Large genomic rearrangements (LGR) in BRCA1 consisting of deletions/duplications of one or several exons have been found throughout the gene with a large proportion occurring in the 5' region from the promoter to exon 2. The aim of this study was to better characterize those LGR in French high-risk breast/ovarian cancer families.
Methods: DNA from 20 families with one apparent duplication and nine deletions was analyzed with a dedicated comparative genomic hybridization (CGH) array, high-resolution BRCA1 Genomic Morse Codes analysis and Sanger sequencing.
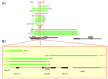
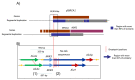
Results: The apparent duplication was in fact a tandem triplication of exons 1 and 2 and part of intron 2 of BRCA1, fully characterized here for the first time. We calculated a causality score with the multifactorial model from data obtained from six families, classifying this variant as benign. Among the nine deletions detected in this region, eight have never been identified. The breakpoints fell in six recurrent regions and could confirm some specific conformation of the chromatin.
Conclusions: Taken together, our results firmly establish that the BRCA1 5' region is a frequent site of different LGRs and highlight the importance of the segmental duplication and Alu sequences, particularly the very high homologous region, in the mechanism of a recombination event. This also confirmed that those events are not systematically deleterious.
Keywords: BRCA1 gene; large duplication; large genomic rearrangement; triplication; variant of unknown significance.
Conflict of interest statement
M.-H.S. is a coinventor of the LST method for detecting inactivation of the HR pathway (BRCA1/2) in human tumors, patent numbers 20170260588 and 20150140122, under exclusive licensing with Myriad Genetics. All other authors declare no conflict of interest.
Figures



References
-
- Caputo: S., Benboudjema L., Sinilnikova O., Rouleau E., Béroud C., Lidereau R. Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40:D992–D1002. doi: 10.1093/nar/gkr1160. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

