Tracking HIV-1-Infected Cell Clones Using Integration Site-Specific qPCR
- PMID: 34202310
- PMCID: PMC8310066
- DOI: 10.3390/v13071235
Tracking HIV-1-Infected Cell Clones Using Integration Site-Specific qPCR
Abstract
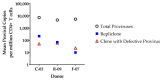
Efforts to cure HIV-1 infection require better quantification of the HIV-1 reservoir, particularly the clones of cells harboring replication-competent (intact) proviruses, termed repliclones. The digital droplet PCR assays commonly used to quantify intact proviruses do not differentiate among specific repliclones, thus the dynamics of repliclones are not well defined. The major challenge in tracking repliclones is the relative rarity of the cells carrying specific intact proviruses. To date, detection and accurate quantification of repliclones requires in-depth integration site sequencing. Here, we describe a simplified workflow using integration site-specific qPCR (IS-qPCR) to determine the frequencies of the proviruses integrated in individual repliclones. We designed IS-qPCR to determine the frequencies of repliclones and clones of cells that carry defective proviruses in samples from three donors. Comparing the results of IS-qPCR with deep integration site sequencing data showed that the two methods yielded concordant estimates of clone frequencies (r = 0.838). IS-qPCR is a potentially valuable tool that can be applied to multiple samples and cell types over time to measure the dynamics of individual repliclones and the efficacy of treatments designed to eliminate them.
Keywords: HIV-1 reservoir; HIV-1-infected cell clones; proviral integration sites; repliclones.
Conflict of interest statement
J.W.M. is a consultant to Gilead Sciences and owns share options in Co-Crystal Pharma, Inc. (Bothell, WA, USA) and Infectious Disease Connect, Inc. (Cranberry Township, PA, USA) and shares of Abound Bio, Inc. (Pittsburgh, PA, USA), unrelated to the current work.
Figures



Similar articles
-
New Approaches to Multi-Parametric HIV-1 Genetics Using Multiple Displacement Amplification: Determining the What, How, and Where of the HIV-1 Reservoir.Viruses. 2021 Dec 10;13(12):2475. doi: 10.3390/v13122475. Viruses. 2021. PMID: 34960744 Free PMC article. Review.
-
Sequence Evaluation and Comparative Analysis of Novel Assays for Intact Proviral HIV-1 DNA.J Virol. 2021 Feb 24;95(6):e01986-20. doi: 10.1128/JVI.01986-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361426 Free PMC article.
-
Deep Sequencing Analysis of Individual HIV-1 Proviruses Reveals Frequent Asymmetric Long Terminal Repeats.J Virol. 2022 Jul 13;96(13):e0012222. doi: 10.1128/jvi.00122-22. Epub 2022 Jun 8. J Virol. 2022. PMID: 35674431 Free PMC article.
-
Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained.Retrovirology. 2018 Feb 17;15(1):22. doi: 10.1186/s12977-018-0396-3. Retrovirology. 2018. PMID: 29452580 Free PMC article. Review.
-
Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections.Viruses. 2020 Jan 25;12(2):136. doi: 10.3390/v12020136. Viruses. 2020. PMID: 31991737 Free PMC article.
Cited by
-
HIV infected CD4+ T cell clones are more stable than uninfected clones during long-term antiretroviral therapy.PLoS Pathog. 2022 Aug 31;18(8):e1010726. doi: 10.1371/journal.ppat.1010726. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 36044447 Free PMC article.
-
CD4+ T cells with latent HIV-1 have reduced proliferative responses to T cell receptor stimulation.J Exp Med. 2024 Mar 4;221(3):e20231511. doi: 10.1084/jem.20231511. Epub 2024 Jan 25. J Exp Med. 2024. PMID: 38270554 Free PMC article.
-
The largest HIV-1-infected T cell clones in children on long-term combination antiretroviral therapy contain solo LTRs.mBio. 2023 Aug 31;14(4):e0111623. doi: 10.1128/mbio.01116-23. Epub 2023 Aug 2. mBio. 2023. PMID: 37530525 Free PMC article.
-
Distinguishable topology of the task-evoked functional genome networks in HIV-1 reservoirs.iScience. 2024 Oct 21;27(11):111222. doi: 10.1016/j.isci.2024.111222. eCollection 2024 Nov 15. iScience. 2024. PMID: 39559761 Free PMC article.
-
Experimental models for HIV latency and molecular tools for reservoir quantification-an update.Clin Microbiol Rev. 2023 Dec 20;36(4):e0001323. doi: 10.1128/cmr.00013-23. Epub 2023 Nov 15. Clin Microbiol Rev. 2023. PMID: 37966222 Free PMC article. Review.
References
-
- Palella F.J., Delaney K.M., Moorman A.C., Loveless M.O., Fuhrer J., Satten G.A., Aschman D.J., Holmberg S.D., Investigators T.H.O.S. Declining Morbidity and Mortality among Patients with Advanced Human Immunodeficiency Virus Infection. N. Engl. J. Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. - DOI - PubMed
-
- Coffin J.M., Bale M.J., Wells D., Guo S., Luke B., Zerbato J.M., Sobolewski M.D., Sia T., Shao W., Wu X., et al. Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLoS Pathog. 2021;17:e1009141. doi: 10.1371/journal.ppat.1009141. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

