The Double Mutation DSG2-p.S363X and TBX20-p.D278X Is Associated with Left Ventricular Non-Compaction Cardiomyopathy: Case Report
- PMID: 34202524
- PMCID: PMC8268202
- DOI: 10.3390/ijms22136775
The Double Mutation DSG2-p.S363X and TBX20-p.D278X Is Associated with Left Ventricular Non-Compaction Cardiomyopathy: Case Report
Abstract
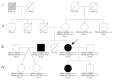
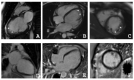
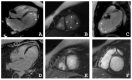
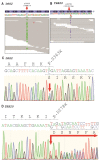
Left ventricular non-compaction cardiomyopathy (LVNC) is a rare heart disease, with or without left ventricular dysfunction, which is characterized by a two-layer structure of the myocardium and an increased number of trabeculae. The study of familial forms of LVNC is helpful for risk prediction and genetic counseling of relatives. Here, we present a family consisting of three members with LVNC. Using a next-generation sequencing approach a combination of two (likely) pathogenic nonsense mutations DSG2-p.S363X and TBX20-p.D278X was identified in all three patients. TBX20 encodes the cardiac T-box transcription factor 20. DSG2 encodes desmoglein-2, which is part of the cardiac desmosomes and belongs to the cadherin family. Since the identified nonsense variant (DSG2-p.S363X) is localized in the extracellular domain of DSG2, we performed in vitro cell transfection experiments. These experiments revealed the absence of truncated DSG2 at the plasma membrane, supporting the pathogenic relevance of DSG2-p.S363X. In conclusion, we suggest that in the future, these findings might be helpful for genetic screening and counseling of patients with LVNC.
Keywords: DSG2; TBX20; cardiomyopathy; cardiovascular genetics; desmoglein-2; desmosomes; dilated cardiomyopathy; left ventricular non-compaction cardiomyopathy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Myasnikov R.P., Kulikova O.V., Meshkov A.N., Kiseleva A.V., Shumarina A.O., Koretskiy S.N., Zharikova A.A., Divashuk M.G., Kharlap M.S., Serduk S.E., et al. New variant of MYH7 gene nucleotide sequence in familial non-compaction cardiomyopathy with benign course. Ration. Pharmacother. Cardiol. 2020;16:383–391. doi: 10.20996/1819-6446-2020-06-01. - DOI
-
- Myasnikov R.P., Blagova O.V., Kulikova O.V., Mershina E.A., Kharlap M.S., Andreenko E.Y., Koretsky S.N., Serdyuk S.E., Bazaeva E.V., Virabova I.A., et al. The specifics of noncompacted cardiomyopathy manifestation. Cardiovasc. Ther. Prev. 2015;14:78. doi: 10.15829/1728-8800-2015-5-78-82. - DOI
-
- Kulikova O.V., Myasnikov R.P., Mershina E.A., Pilus P.S., Koretskiy S.N., Meshkov A.N., Kiseleva A.V., Kharlap M.S., Sinitsyn V.E., Sdvigova N.A., et al. The familial form of non-compaction cardiomyopathy: Types of myocardial remodeling, clinical course. Multicenter register results. Ther. Arch. 2021;94 doi: 10.26442/00403660.2021.04.200677. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

