Sp1-Induced FNBP1 Drives Rigorous 3D Cell Motility in EMT-Type Gastric Cancer Cells
- PMID: 34202606
- PMCID: PMC8267707
- DOI: 10.3390/ijms22136784
Sp1-Induced FNBP1 Drives Rigorous 3D Cell Motility in EMT-Type Gastric Cancer Cells
Abstract
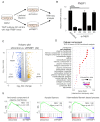
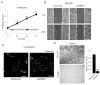
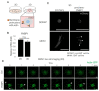
Cancer is heterogeneous among patients, requiring a thorough understanding of molecular subtypes and the establishment of therapeutic strategies based on its behavior. Gastric cancer (GC) is adenocarcinoma with marked heterogeneity leading to different prognoses. As an effort, we previously identified a stem-like subtype, which is prone to metastasis, with the worst prognosis. Here, we propose FNBP1 as a key to high-level cell motility, present only in aggressive GC cells. FNBP1 is also up-regulated in both the GS subtype from the TCGA project and the EMT subtype from the ACRG study, which include high portions of diffuse histologic type. Ablation of FNBP1 in the EMT-type GC cell line brought changes in the cell periphery in transcriptomic analysis. Indeed, loss of FNBP1 resulted in the loss of invasive ability, especially in a three-dimensional culture system. Live imaging indicated active movement of actin in FNBP1-overexpressed cells cultured in an extracellular matrix dome. To find the transcription factor which drives FNBP1 expression in an EMT-type GC cell line, the FNBP1 promoter region and DNA binding motifs were analyzed. Interestingly, the Sp1 motif was abundant in the promoter, and pharmacological inhibition and knockdown of Sp1 down-regulated FNBP1 promoter activity and the transcription level, respectively. Taken together, our results propose Sp1-driven FNBP1 as a key molecule explaining aggressiveness in EMT-type GC cells.
Keywords: EMT; FNBP1; Sp1; cell motility; gastric cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lei Z., Tan I.B., Das K., Deng N., Zouridis H., Pattison S., Chua C., Feng Z., Guan Y.K., Ooi C.H., et al. Identification of molecular subtypes of gastric cancer with different responses to pi3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–565. doi: 10.1053/j.gastro.2013.05.010. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

