Genome-Wide Atlas of Promoter Expression Reveals Contribution of Transcribed Regulatory Elements to Genetic Control of Disuse-Mediated Atrophy of Skeletal Muscle
- PMID: 34203013
- PMCID: PMC8235325
- DOI: 10.3390/biology10060557
Genome-Wide Atlas of Promoter Expression Reveals Contribution of Transcribed Regulatory Elements to Genetic Control of Disuse-Mediated Atrophy of Skeletal Muscle
Abstract
The prevention of muscle atrophy carries with it clinical significance for the control of increased morbidity and mortality following physical inactivity. While major transcriptional events associated with muscle atrophy-recovery processes are the subject of active research on the gene level, the contribution of non-coding regulatory elements and alternative promoter usage is a major source for both the production of alternative protein products and new insights into the activity of transcription factors. We used the cap-analysis of gene expression (CAGE) to create a genome-wide atlas of promoter-level transcription in fast (m. EDL) and slow (m. soleus) muscles in rats that were subjected to hindlimb unloading and subsequent recovery. We found that the genetic regulation of the atrophy-recovery cycle in two types of muscle is mediated by different pathways, including a unique set of non-coding transcribed regulatory elements. We showed that the activation of "shadow" enhancers is tightly linked to specific stages of atrophy and recovery dynamics, with the largest number of specific regulatory elements being transcriptionally active in the muscles on the first day of recovery after a week of disuse. The developed comprehensive database of transcription of regulatory elements will further stimulate research on the gene regulation of muscle homeostasis in mammals.
Keywords: RNA transcription; atrophy; cis-regulatory elements; disuse; enhancers; promoters; rat; skeletal muscles; transcribed non-coding elements of genome; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


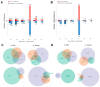
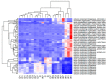
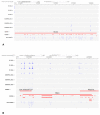

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

