Investigation of Salmonella Phage-Bacteria Infection Profiles: Network Structure Reveals a Gradient of Target-Range from Generalist to Specialist Phage Clones in Nested Subsets
- PMID: 34203492
- PMCID: PMC8310288
- DOI: 10.3390/v13071261
Investigation of Salmonella Phage-Bacteria Infection Profiles: Network Structure Reveals a Gradient of Target-Range from Generalist to Specialist Phage Clones in Nested Subsets
Abstract
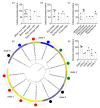
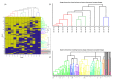
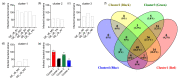
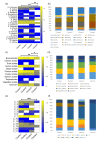
Bacteriophages that lyse Salmonella enterica are potential tools to target and control Salmonella infections. Investigating the host range of Salmonella phages is a key to understand their impact on bacterial ecology, coevolution and inform their use in intervention strategies. Virus-host infection networks have been used to characterize the "predator-prey" interactions between phages and bacteria and provide insights into host range and specificity. Here, we characterize the target-range and infection profiles of 13 Salmonella phage clones against a diverse set of 141 Salmonella strains. The environmental source and taxonomy contributed to the observed infection profiles, and genetically proximal phages shared similar infection profiles. Using in vitro infection data, we analyzed the structure of the Salmonella phage-bacteria infection network. The network has a non-random nested organization and weak modularity suggesting a gradient of target-range from generalist to specialist species with nested subsets, which are also observed within and across the different phage infection profile groups. Our results have implications for our understanding of the coevolutionary mechanisms shaping the ecological interactions between Salmonella phages and their bacterial hosts and can inform strategies for targeting Salmonella enterica with specific phage preparations.
Keywords: bacteria; bacteriophages; evolution; infection; modularity; nestedness; network; salmonella; speciation; virus.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- World Health Organization Salmonella (Non-Typhoidal). Newsroom. [(accessed on 6 April 2021)];2018 Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal)
-
- Centers for Disease Control and Prevention Salmonella. [(accessed on 6 May 2021)]; Available online: https://www.cdc.gov/salmonella/index.html.
-
- Wang X., Biswas S., Paudyal N., Pan H., Li X., Fang W., Yue M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front. Microbiol. 2019;10:985. doi: 10.3389/fmicb.2019.00985. - DOI - PMC - PubMed
-
- Chanishvili N. Literature Review of the Practical Application of Bacteriophage Research. George Eliava Institute of Bacteriophage, Microbiology and Virology; Tbilisi, Georgia: Nova Science Publishers; Hauppauge, NY, USA: 2012. Bacteriology Research Developments, Virology Research Progress.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

