ADCC-Inducing Antibody Trastuzumab and Selection of KIR-HLA Ligand Mismatched Donors Enhance the NK Cell Anti-Breast Cancer Response
- PMID: 34203549
- PMCID: PMC8268223
- DOI: 10.3390/cancers13133232
ADCC-Inducing Antibody Trastuzumab and Selection of KIR-HLA Ligand Mismatched Donors Enhance the NK Cell Anti-Breast Cancer Response
Abstract
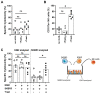
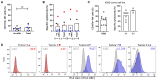
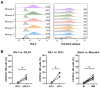
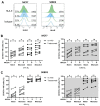
Natural killer (NK)-cell-based immunotherapies are an attractive treatment option for cancer. We previously showed that alloreactive mouse NK cells cured mice of 4T1 breast cancer. However, the tumor microenvironment can inhibit immune responses, and these suppressive factors must be overcome to unfold the NK cells' full anti-tumor potential. Here, we investigated the combination of antibody-dependent cellular cytotoxicity (ADDC) and the selection of KIR-HLA-ligand mismatched NK cells to enhance NK cell anti-breast cancer responses in clinically relevant settings. Donor-derived and IL-2-activated NK cells were co-cultured with patient-derived breast cancer cells or cell lines MCF7 or SKBR3 together with the anti-HER2 antibody trastuzumab. NK cells mediated anti-breast cancer cytotoxicity under normoxic and hypoxic conditions. Under both conditions, trastuzumab vigorously enhanced NK cell degranulation (CD107a) against HER2-overexpressing SKBR3 cells, but we observed a discrepancy between highly degranulating NK cells and a rather modest increase in cytotoxicity of SKBR3. Against patient-derived breast cancer cells, the anti-tumor efficacy was rather limited, and HLA class I expression seemed to contribute to inhibited NK cell functionality. KIR-ligand-mismatched NK cells degranulated stronger compared to the matched NK cells, further highlighting the role of HLA. In summary, trastuzumab and KIR-ligand-mismatched NK cells could be two strategies to potently enhance NK cell responses to breast cancer.
Keywords: HLA class I; alloreactive donor NK cells; antibody-dependent cellular cytotoxicity; breast cancer; tumor microenvironment.
Conflict of interest statement
G.M.J.B. is Chief Executive Officer/Chief Medical Officer/Cofounder of CiMaas, BV, Maastricht, The Netherlands. CiMaas is producing an ex vivo expanded NK cell product that will be used to treat myeloma patients.
Figures


References
-
- Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. Arch. Pathol. Lab. Med. 2018;142:1364–1382. doi: 10.5858/arpa.2018-0902-SA. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

