From Sampling to Sequencing: A Liquid Biopsy Pre-Analytic Workflow to Maximize Multi-Layer Genomic Information from a Single Tube
- PMID: 34203921
- PMCID: PMC8232701
- DOI: 10.3390/cancers13123002
From Sampling to Sequencing: A Liquid Biopsy Pre-Analytic Workflow to Maximize Multi-Layer Genomic Information from a Single Tube
Abstract
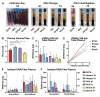
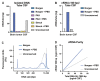
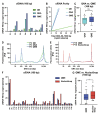
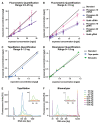
Liquid biopsies hold great promise for the management of cancer. Reliable liquid biopsy data depend on stable and reproducible pre-analytical protocols that comply with quality measures, irrespective of the sampling and processing site. We established a workflow for plasma preservation, followed by processing, cell-free nucleic acid isolation, quantification, and enrichment of potentially tumor-derived cell-free DNA and RNA. Employing the same input material for a direct comparison of different kits and protocols allowed us to formulate unbiased recommendations for sample collection, storage, and processing. The presented workflow integrates the stabilization in Norgen, PAX, or Streck tubes and subsequent parallel isolation of cell-free DNA and RNA with NucleoSnap and NucleoSpin. Qubit, Bioanalyzer, and TapeStation quantification and quality control steps were optimized for minimal sample use and high sensitivity and reproducibility. We show the efficiency of the proposed workflow by successful droplet digital PCR amplification of both cell-free DNA and RNA and by detection of tumor-specific alterations in low-coverage whole-genome sequencing and DNA methylation profiling of plasma-derived cell-free DNA. For the first time, we demonstrated successful parallel extraction of cell-free DNA and RNA from plasma samples. This workflow paves the road towards multi-layer genomic analysis from one single liquid biopsy sample.
Keywords: CSF; blood preservation tubes; cancer; cell-free DNA; cell-free RNA; droplet digital PCR; liquid biopsy; low-coverage whole-genome sequencing; pre-analytics; size selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
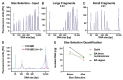
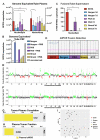
References
-
- Sorenson G.D., Pribish D.M., Valone F.H., Memoli V.A., Bzik D.J., Yao S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomark. Prev. 1994;3:67–71. - PubMed
-
- Olsson E., Winter C., George A., Chen Y., Howlin J., Tang M.H., Dahlgren M., Schulz R., Grabau D., van Westen D., et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 2015;7:1034–1047. doi: 10.15252/emmm.201404913. - DOI - PMC - PubMed
-
- Parkinson C.A., Gale D., Piskorz A.M., Biggs H., Hodgkin C., Addley H., Freeman S., Moyle P., Sala E., Sayal K., et al. Exploratory analysis of tp53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: A retrospective study. PLoS Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

