Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era
- PMID: 34204069
- PMCID: PMC8229000
- DOI: 10.3390/biology10060504
Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era
Abstract
Synthetic gene circuits are made of DNA sequences, referred to as transcription units, that communicate by exchanging proteins or RNA molecules. Proteins are, mostly, transcription factors that bind promoter sequences to modulate the expression of other molecules. Promoters are, therefore, key components in genetic circuits. In this review, we focus our attention on the construction of artificial promoters for the yeast S. cerevisiae, a popular chassis for gene circuits. We describe the initial techniques and achievements in promoter engineering that predated the start of the Synthetic Biology epoch of about 20 years. We present the main applications of synthetic promoters built via different methods and discuss the latest innovations in the wet-lab engineering of novel promoter sequences.
Keywords: Saccharomyces cerevisiae; gene expression; promoter; synthetic biology; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

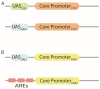
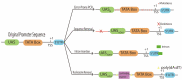

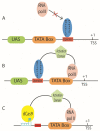
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

