TOMM40 RNA Transcription in Alzheimer's Disease Brain and Its Implication in Mitochondrial Dysfunction
- PMID: 34204109
- PMCID: PMC8226536
- DOI: 10.3390/genes12060871
TOMM40 RNA Transcription in Alzheimer's Disease Brain and Its Implication in Mitochondrial Dysfunction
Abstract
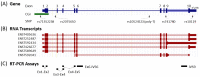
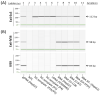
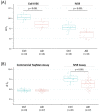
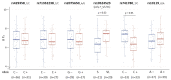
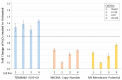
Increasing evidence suggests that the Translocase of Outer Mitochondria Membrane 40 (TOMM40) gene may contribute to the risk of Alzheimer's disease (AD). Currently, there is no consensus as to whether TOMM40 expression is up- or down-regulated in AD brains, hindering a clear interpretation of TOMM40's role in this disease. The aim of this study was to determine if TOMM40 RNA levels differ between AD and control brains. We applied RT-qPCR to study TOMM40 transcription in human postmortem brain (PMB) and assessed associations of these RNA levels with genetic variants in APOE and TOMM40. We also compared TOMM40 RNA levels with mitochondrial functions in human cell lines. Initially, we found that the human genome carries multiple TOMM40 pseudogenes capable of producing highly homologous RNAs that can obscure precise TOMM40 RNA measurements. To circumvent this obstacle, we developed a novel RNA expression assay targeting the primary transcript of TOMM40. Using this assay, we showed that TOMM40 RNA was upregulated in AD PMB. Additionally, elevated TOMM40 RNA levels were associated with decreases in mitochondrial DNA copy number and mitochondrial membrane potential in oxidative stress-challenged cells. Overall, differential transcription of TOMM40 RNA in the brain is associated with AD and could be an indicator of mitochondrial dysfunction.
Keywords: Alzheimer’s disease; RNA transcription; TOMM40 gene; mitochondrial dysfunction; pseudogene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. - DOI - PMC - PubMed
-
- Beecham G.W., Hamilton K., Naj A.C., Martin E.R., Huentelman M., Myers A.J., Corneveaux J.J., Hardy J., Vonsattel J.P., Younkin S.G., et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606. doi: 10.1371/journal.pgen.1004606. - DOI - PMC - PubMed
-
- Yashin A.I., Fang F., Kovtun M., Wu D., Duan M., Arbeev K., Akushevich I., Kulminski A., Culminskaya I., Zhbannikov I., et al. Hidden heterogeneity in Alzheimer’s disease: Insights from genetic association studies and other analyses. Exp. Gerontol. 2018;107:148–160. doi: 10.1016/j.exger.2017.10.020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

