Genotypic and Phenotypic Characterization of Highly Alkaline-Resistant Carnobacterium maltaromaticum V-Type ATPase from the Dairy Product Based on Comparative Genomics
- PMID: 34204143
- PMCID: PMC8229585
- DOI: 10.3390/microorganisms9061233
Genotypic and Phenotypic Characterization of Highly Alkaline-Resistant Carnobacterium maltaromaticum V-Type ATPase from the Dairy Product Based on Comparative Genomics
Abstract
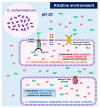
Although Carnobacterium maltaromaticum derived from dairy products has been used as a lactic acid bacterium industrially, several studies have reported potential pathogenicity and disease outbreaks. Because strains derived from diseased fish and dairy products are considered potentially virulent and beneficial, respectively, their genotypic and phenotypic characteristics have attracted considerable attention. A genome-wide comparison of 30 genome sequences (13, 3, and 14 strains from diseased aquatic animals, dairy products, and processed food, respectively) was carried out. Additionally, one dairy and two nondairy strains were incubated in nutrient-rich (diluted liquid media) and nutrient-deficient environments (PBS) at pH 10 to compare their alkaline resistance in accordance with different nutritional environments by measuring their optical density and viable bacterial cell counts. Interestingly, only dairy strains carried 11 shared accessory genes, and 8 genes were strongly involved in the V-type ATPase gene cluster. Given that V-type ATPase contributes to resistance to alkaline pH and salts using proton motive force generated via sodium translocation across the membrane, C. maltaromaticum with a V-type ATPase might use nutrients in food under high pH. Indeed, the dairy strain carrying the V-type ATPase exhibited the highest alkaline resistance only in the nutrient-rich environment with significant upregulation of V-type ATPase expression. These results suggest that the gene cluster of V-type ATPase and increased alkaline resistance of dairy strains facilitate adaptation in the long-term ripening of alkaline dairy products.
Keywords: V-type ATPase; alkaline resistance; bioinformatics; food quality; genomics; lactic acid bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Carnobacterium divergens and Carnobacterium maltaromaticum as spoilers or protective cultures in meat and seafood: phenotypic and genotypic characterization.Syst Appl Microbiol. 2005 Mar;28(2):151-64. doi: 10.1016/j.syapm.2004.12.001. Syst Appl Microbiol. 2005. PMID: 15830808
-
Genome-wide comparison of Carnobacterium maltaromaticum derived from diseased fish harbouring important virulence-related genes.J Fish Dis. 2020 Sep;43(9):1029-1037. doi: 10.1111/jfd.13208. Epub 2020 Jul 6. J Fish Dis. 2020. PMID: 32627213
-
Multilocus sequence typing of Carnobacterium maltaromaticum strains associated with fish disease and dairy products.J Appl Microbiol. 2019 Feb;126(2):377-387. doi: 10.1111/jam.14127. Epub 2018 Nov 22. J Appl Microbiol. 2019. PMID: 30307684
-
Carnobacterium maltaromaticum: identification, isolation tools, ecology and technological aspects in dairy products.Food Microbiol. 2010 Aug;27(5):573-9. doi: 10.1016/j.fm.2010.03.019. Epub 2010 Apr 8. Food Microbiol. 2010. PMID: 20510773 Review.
-
Safety and nutritional assessment of GM plants and derived food and feed: the role of animal feeding trials.Food Chem Toxicol. 2008 Mar;46 Suppl 1:S2-70. doi: 10.1016/j.fct.2008.02.008. Epub 2008 Feb 13. Food Chem Toxicol. 2008. PMID: 18328408 Review.
Cited by
-
A genome-wide association study of the occurrence of genetic variations in Edwardsiella piscicida, Vibrio harveyi, and Streptococcus parauberis under stressed environments.J Fish Dis. 2022 Sep;45(9):1373-1388. doi: 10.1111/jfd.13668. Epub 2022 Jun 23. J Fish Dis. 2022. PMID: 35735095 Free PMC article.
-
The potential regulatory role of mannose phosphotransferase system EII in alkaline resistance of Enterococcus faecalis.J Oral Microbiol. 2025 Apr 6;17(1):2487944. doi: 10.1080/20002297.2025.2487944. eCollection 2025. J Oral Microbiol. 2025. PMID: 40206098 Free PMC article.
-
Genomic and Transcriptomic Diversification of Flagellin Genes Provides Insight into Environmental Adaptation and Phylogeographic Characteristics in Aeromonas hydrophila.Microb Ecol. 2024 May 2;87(1):65. doi: 10.1007/s00248-024-02373-4. Microb Ecol. 2024. PMID: 38695873 Free PMC article.
References
-
- Gómez N.C., Ramiro J.M., Quecan B.X., de Melo Franco B.D. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016;7:863. doi: 10.3389/fmicb.2016.00863. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

