Klebsiella pneumoniae Lipopolysaccharides Serotype O2afg Induce Poor Inflammatory Immune Responses Ex Vivo
- PMID: 34204279
- PMCID: PMC8234205
- DOI: 10.3390/microorganisms9061317
Klebsiella pneumoniae Lipopolysaccharides Serotype O2afg Induce Poor Inflammatory Immune Responses Ex Vivo
Abstract
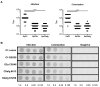
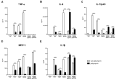
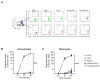
Currently, Klebsiella pneumoniae is a pathogen of clinical relevance due to its plastic ability of acquiring resistance genes to multiple antibiotics. During K. pneumoniae infections, lipopolysaccharides (LPS) play an ambiguous role as they both activate immune responses but can also play a role in immune evasion. The LPS O2a and LPS O2afg serotypes are prevalent in most multidrug resistant K. pneumoniae strains. Thus, we sought to understand if those two particular LPS serotypes were involved in a mechanism of immune evasion. We have extracted LPS (serotypes O1, O2a and O2afg) from K. pneumoniae strains and, using human monocytes ex vivo, we assessed the ability of those LPS antigens to induce the production of pro-inflammatory cytokines and chemokines. We observed that, when human monocytes are incubated with LPS serotypes O1, O2a or O2afg strains, O2afg and, to a lesser extent, O2a but not O1 failed to elicit the production of pro-inflammatory cytokines and chemokines, which suggests a role in immune evasion. Our preliminary data also shows that nuclear translocation of NF-κB, a process which regulates an immune response against infections, occurs in monocytes incubated with LPS O1 and, to a smaller extent, with LPS O2a, but not with the LPS serotype O2afg. Our results indicate that multidrug resistant K. pneumoniae expressing LPS O2afg serotypes avoid an initial inflammatory immune response and, consequently, are able to systematically spread inside the host unharmed, which results in the several pathologies associated with this bacterium.
Keywords: Klebsiella pneumoniae; NF-κB; antimicrobial resistance; immune evasion; lipopolysaccharides; nosocomial infection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Figures


References
-
- WHO WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. [(accessed on 20 March 2017)]; Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-n...
-
- Dalton K.R., Rock C., Carroll K.C., Davis M.F. One Health in hospitals: How understanding the dynamics of people, animals, and the hospital built-environment can be used to better inform interventions for antimicrobial-resistant gram-positive infections. Antimicrob. Resist. Infect. Control. 2020;9:78. doi: 10.1186/s13756-020-00737-2. - DOI - PMC - PubMed
-
- Temkin E., Fallach N., Almagor J., Gladstone B.P., Tacconelli E., Carmeli Y. Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: A modelling study. Lancet Glob. Health. 2018;6:e969–e979. doi: 10.1016/S2214-109X(18)30278-X. - DOI - PubMed
LinkOut - more resources
Full Text Sources

