The Role of Coronavirus RNA-Processing Enzymes in Innate Immune Evasion
- PMID: 34204549
- PMCID: PMC8235370
- DOI: 10.3390/life11060571
The Role of Coronavirus RNA-Processing Enzymes in Innate Immune Evasion
Abstract
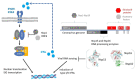
Viral RNA sensing triggers innate antiviral responses in humans by stimulating signaling pathways that include crucial antiviral genes such as interferon. RNA viruses have evolved strategies to inhibit or escape these mechanisms. Coronaviruses use multiple enzymes to synthesize, modify, and process their genomic RNA and sub-genomic RNAs. These include Nsp15 and Nsp16, whose respective roles in RNA capping and dsRNA degradation play a crucial role in coronavirus escape from immune surveillance. Evolutionary studies on coronaviruses demonstrate that genome expansion in Nidoviruses was promoted by the emergence of Nsp14-ExoN activity and led to the acquisition of Nsp15- and Nsp16-RNA-processing activities. In this review, we discuss the main RNA-sensing mechanisms in humans as well as recent structural, functional, and evolutionary insights into coronavirus Nsp15 and Nsp16 with a view to potential antiviral strategies.
Keywords: SARS-CoV-2; coronavirus; immune evasion; innate immunity; viral RNA sensing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Xie J., Wu W., Li S., Hu Y., Hu M., Li J., Yang Y., Huang T., Zheng K., Wang Y., et al. Clinical characteristics and outcomes of critically ill patients with novel coronavirus infectious disease (COVID-19) in China: A retrospective multicenter study. Intensive Care Med. 2020;46:1863–1872. doi: 10.1007/s00134-020-06211-2. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

