The Exon Junction Complex Core Represses Cancer-Specific Mature mRNA Re-splicing: A Potential Key Role in Terminating Splicing
- PMID: 34204574
- PMCID: PMC8234774
- DOI: 10.3390/ijms22126519
The Exon Junction Complex Core Represses Cancer-Specific Mature mRNA Re-splicing: A Potential Key Role in Terminating Splicing
Abstract
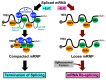
Using TSG101 pre-mRNA, we previously discovered cancer-specific re-splicing of mature mRNA that generates aberrant transcripts/proteins. The fact that mRNA is aberrantly re-spliced in various cancer cells implies there must be an important mechanism to prevent deleterious re-splicing on the spliced mRNA in normal cells. We thus postulated that mRNA re-splicing is controlled by specific repressors, and we searched for repressor candidates by siRNA-based screening for mRNA re-splicing activity. We found that knock-down of EIF4A3, which is a core component of the exon junction complex (EJC), significantly promoted mRNA re-splicing. Remarkably, we could recapitulate cancer-specific mRNA re-splicing in normal cells by knock-down of any of the core EJC proteins, EIF4A3, MAGOH, or RBM8A (Y14), implicating the EJC core as the repressor of mRNA re-splicing often observed in cancer cells. We propose that the EJC core is a critical mRNA quality control factor to prevent over-splicing of mature mRNA.
Keywords: EIF4A3; EJC; MAGOH; RBM8A (Y14); TSG101; mRNA re-splicing; pre-mRNA splicing.
Conflict of interest statement
Y.O. was delegated to the Mayeda Laboratory as a visiting student from Nippon Shinyaku Co., Ltd., and he received financial support from the company. The other authors have no financial conflicts of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

